8QYJ
 
 | | Human 20S proteasome assembly structure 1 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome assembly chaperone 3, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYO
 
 | | Human proteasome 20S core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QZ9
 
 | | Human 20S proteasome assembly intermediate structure 4 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYL
 
 | | Human 20S proteasome assembly intermediate structure 2 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYM
 
 | | Human 20S proteasome assembly intermediate structure 3 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYS
 
 | | Human preholo proteasome 20S core particle | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYN
 
 | | Human 20S proteasome assembly intermediate structure 5 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3IVQ
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-CiSBC2 | | Descriptor: | CiSBC2, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVV
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-PucSBC1_pep1 | | Descriptor: | PucSBC1, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3IVB
 
 | | Structures of SPOP-Substrate Complexes: Insights into Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-MacroH2ASBCpep1 | | Descriptor: | CHLORIDE ION, Core histone macro-H2A.1, Speckle-type POZ protein, ... | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure 9
To be Published
|
|
1FQV
 
 | | Insights into scf ubiquitin ligases from the structure of the skp1-skp2 complex | | Descriptor: | SKP1, SKP2 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-06 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
1FS2
 
 | | INSIGHTS INTO SCF UBIQUITIN LIGASES FROM THE STRUCTURE OF THE SKP1-SKP2 COMPLEX | | Descriptor: | SKP1, SKP2 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R.E, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
1FS1
 
 | | INSIGHTS INTO SCF UBIQUITIN LIGASES FROM THE STRUCTURE OF THE SKP1-SKP2 COMPLEX | | Descriptor: | CYCLIN A/CDK2-ASSOCIATED P19, CYCLIN A/CDK2-ASSOCIATED P45 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R.E, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
7ADO
 
 | | Cryo-EM structure of human ER membrane protein complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7WUG
 
 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | Descriptor: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Cheng, J.D, Schulman, B.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
6TTU
 
 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
6BG5
 
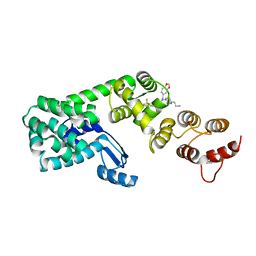 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
2AST
 
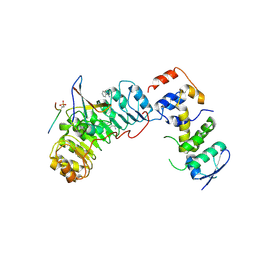 | | Crystal structure of Skp1-Skp2-Cks1 in complex with a p27 peptide | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
7Q51
 
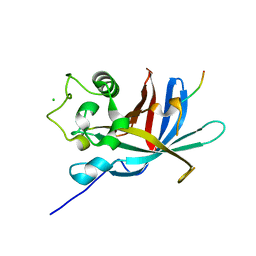 | | yeast Gid10 bound to a Phe/N-peptide | | Descriptor: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
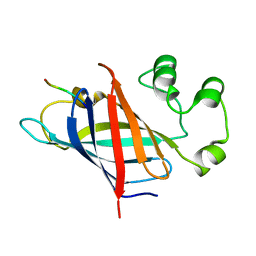 | | human Gid4 bound to a Phe/N-peptide | | Descriptor: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | Authors: | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7QQY
 
 | | yeast Gid10 bound to Art2 Pro/N-degron | | Descriptor: | CHLORIDE ION, ECM21, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | A GID E3 ligase assembly ubiquitinates an Rsp5 E3 adaptor and regulates plasma membrane transporters.
Embo Rep., 23, 2022
|
|
7Q4Y
 
 | |
4YII
 
 | |
7Z8B
 
 | |
6SWY
 
 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
