3MPG
 
 | |
1T5A
 
 | | Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Dombrauckas, J.D, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2004-05-03 | | Release date: | 2005-07-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis.
Biochemistry, 44, 2005
|
|
2IDM
 
 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
4RNA
 
 | | Crystal structure of PLpro from Middle East Respiratory Syndrome (MERS) coronavirus | | Descriptor: | PHOSPHATE ION, ZINC ION, papain-like protease | | Authors: | Lei, H, Santarsiero, B.D, Lee, H, Johnson, M.E. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV.
Acs Chem.Biol., 10, 2015
|
|
4YIW
 
 | | DIHYDROOROTASE FROM BACILLUS ANTHRACIS WITH SUBSTRATE BOUND | | Descriptor: | Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ZINC ION | | Authors: | Lei, H, Santarsiero, B.D, Rice, A.J, Lee, H, Johnson, M.E. | | Deposit date: | 2015-03-02 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ca-asp bound X-ray structure and inhibition of Bacillus anthracis dihydroorotase (DHOase).
Bioorg.Med.Chem., 24, 2016
|
|
7L00
 
 | |
2ALV
 
 | | X-ray structural analysis of SARS coronavirus 3CL proteinase in complex with designed anti-viral inhibitors | | Descriptor: | N-((3S,6R)-6-((S,E)-4-ETHOXYCARBONYL-1-((S)-2-OXOPYRROLIDIN-3-YL)BUT-3-EN-2-YLCARBAMOYL)-2,9-DIMETHYL-4-OXODEC-8-EN-3-YL)-5-METHYLISOXAZOLE-3-CARBOXAMIDE, Replicase polyprotein 1ab | | Authors: | Ghosh, A.K, Xi, K, Ratia, K, Santarsiero, B.D, Fu, W, Harcourt, B.H, Rota, P.A, Baker, S.C, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2005-08-08 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors.
J.Med.Chem., 48, 2005
|
|
4DLK
 
 | | Crystal Structure of ATP-Ca++ complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2012-02-06 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2ZC1
 
 | | Organophosphorus Hydrolase from Deinococcus radiodurans | | Descriptor: | BROMIDE ION, COBALT (II) ION, Phosphotriesterase | | Authors: | Larsen, S.D, Hawwa, R, Ratia, K, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2007-11-02 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structural Insights into a Phosphotriesterase
to be published
|
|
1RTP
 
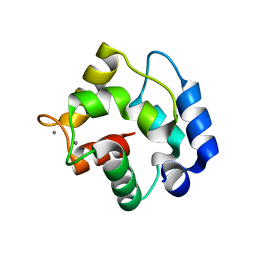 | | REFINED X-RAY STRUCTURE OF RAT PARVALBUMIN, A MAMMALIAN ALPHA-LINEAGE PARVALBUMIN, AT 2.0 A RESOLUTION | | Descriptor: | ALPHA-PARVALBUMIN, CALCIUM ION | | Authors: | Mcphalen, C.A, Sielecki, A.R, Santarsiero, B.D, James, M.N.G. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of rat parvalbumin, a mammalian alpha-lineage parvalbumin, at 2.0 A resolution.
J.Mol.Biol., 235, 1994
|
|
3V4S
 
 | | Crystal Structure of ADP-ATP complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-12-15 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2ID9
 
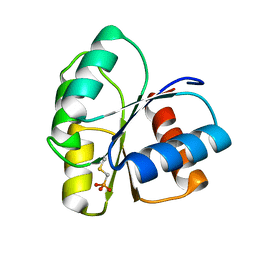 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2ID7
 
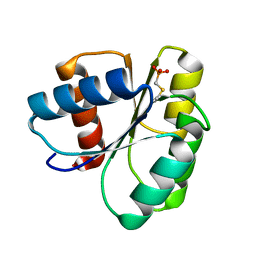 | | 1.75 A Structure of T87I Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
3UIC
 
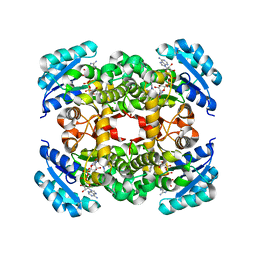 | | Crystal Structure of FabI, an Enoyl Reductase from F. tularensis, in complex with a Novel and Potent Inhibitor | | Descriptor: | 1-(3,4-dichlorobenzyl)-5,6-dimethyl-1H-benzimidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2011-11-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and enzymatic analyses reveal the binding mode of a novel series of Francisella tularensis enoyl reductase (FabI) inhibitors.
J.Med.Chem., 55, 2012
|
|
3F4C
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10, with glycerol bound | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
3F4D
 
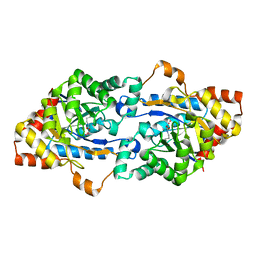 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10 | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
1B71
 
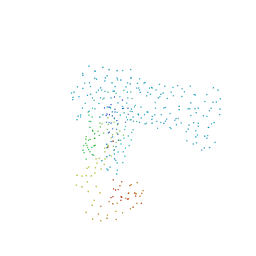 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, PROTEIN (RUBRERYTHRIN), ZINC ION | | Authors: | Sieker, L.C, Holmes, M, Le Trong, I, Turley, S, Santarsiero, B.D, Liu, M.-Y, Legall, J, Stenkamp, R.E. | | Deposit date: | 1999-01-26 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative metal-binding sites in rubrerythrin.
Nat.Struct.Biol., 6, 1999
|
|
2FE8
 
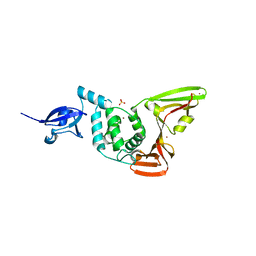 | | SARS coronavirus papain-like protease: structure of a viral deubiquitinating enzyme | | Descriptor: | BROMIDE ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Ratia, K, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2005-12-15 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DWU
 
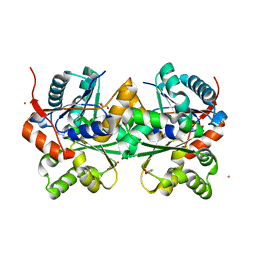 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | Authors: | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2006-08-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
4IQL
 
 | | Crystal Structure of Porphyromonas gingivalis Enoyl-ACP Reductase II (FabK) with cofactors NADPH and FMN | | Descriptor: | Enoyl-(Acyl-carrier-protein) reductase II, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Hevener, K.E, Santarsiero, B.D, Su, P.-C, Boci, T, Truong, K, Johnson, M.E, Mehboob, S. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural characterization of Porphyromonas gingivalis enoyl-ACP reductase II (FabK).
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4J3F
 
 | | Crystal Structure of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold. | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-5,6,7,8-tetrahydro-1H-naphtho[2,3-d]imidazole, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4J4T
 
 | | Crystal Structure of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold | | Descriptor: | 1-(1,3-benzodioxol-5-ylmethyl)-5,6,7,8-tetrahydro-1H-naphtho[2,3-d]imidazole, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2GZM
 
 | | Crystal Structure of the Glutamate Racemase from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | May, M, Santarsiero, B.D, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design.
J.Mol.Biol., 371, 2007
|
|
3FCT
 
 | | MATURE METAL CHELATASE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Romesberg, F.E, Santarsiero, B.D, Barnes, D, Yin, J, Spiller, B, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic evidence for strain in biological catalysis.
Biochemistry, 37, 1998
|
|
4J1N
 
 | | Crystal structures of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-1,5,6,7-tetrahydroindeno[5,6-d]imidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
