5V0N
 
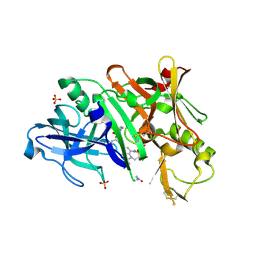 | | BACE1 in complex with inhibitor 5g | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1S,2S)-1-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-hydroxy-3-phenylpropan-2-yl}-7-ethyl-1,3,3-trimethyl-2,2-dioxo-1,2,3,4-tetrahydro-2lambda~6~-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide, ... | | Authors: | Mesecar, A, Ghosh, A, Yen, Y.-C. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Design, synthesis, and X-ray structural studies of BACE-1 inhibitors containing substituted 2-oxopiperazines as P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1AI2
 
 | | ISOCITRATE DEHYDROGENASE COMPLEXED WITH ISOCITRATE, NADP+, AND CALCIUM (FLASH-COOLED) | | Descriptor: | ISOCITRATE CALCIUM COMPLEX, ISOCITRATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Stoddard, B.L, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1997-04-30 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
Science, 277, 1997
|
|
1AI3
 
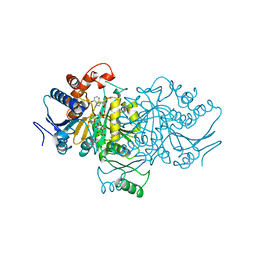 | | ORBITAL STEERING IN THE CATALYTIC POWER OF ENZYMES: SMALL STRUCTURAL CHANGES WITH LARGE CATALYTIC CONSEQUENCES | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
Science, 277, 1997
|
|
1A3X
 
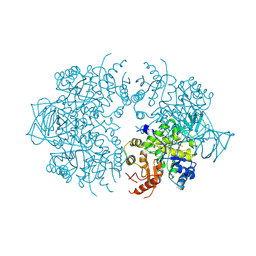 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH PG, MN2+ AND K+ | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
1A3W
 
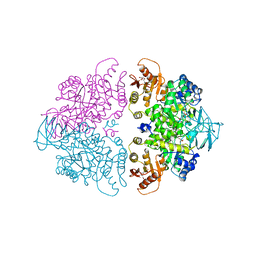 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH FBP, PG, MN2+ AND K+ | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
1BL5
 
 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | Descriptor: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1998-07-23 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
3V3M
 
 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
1TX7
 
 | | Bovine Trypsin complexed with p-amidinophenylmethylphosphinic acid (AMPA) | | Descriptor: | (4-CARBAMIMIDOYLPHENYL)-METHYL-PHOSPHINIC ACID, CALCIUM ION, Trypsinogen | | Authors: | Cui, J, Marankan, F, Fu, W, Crich, D, Mesecar, A, Johnson, M.E. | | Deposit date: | 2004-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An oxyanion-hole selective serine protease inhibitor in complex with trypsin.
Bioorg.Med.Chem., 10, 2002
|
|
6XR3
 
 | |
6WNP
 
 | |
3GTX
 
 | | D71G/E101G mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GTI
 
 | | D71G/E101G/M234L mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase, SODIUM ION | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GTH
 
 | | D71G/E101G/M234I mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, FORMIC ACID, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU1
 
 | | Y97W mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU2
 
 | | Y97L/G100-/E101- mutant in organophosphorus hydrolase | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GU9
 
 | | R228A mutation in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-28 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3GTF
 
 | | D71G/E101G/V235L mutant in organophosphorus hydrolase from Deinococcus radiodurans | | Descriptor: | COBALT (II) ION, Organophosphorus hydrolase | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
3HTW
 
 | | Organophosphorus hydrolase from Deinococcus radiodurans with cacodylate bound | | Descriptor: | CACODYLATE ION, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Hawwa, R, Larsen, S, Ratia, K, Mesecar, A. | | Deposit date: | 2009-06-12 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from Deinococcus radiodurans.
J.Mol.Biol., 393, 2009
|
|
4YO9
 
 | | HKU4 3CLpro unbound structure | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, ACETATE ION, ... | | Authors: | St John, S.E, Mesecar, A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
4YOJ
 
 | | HKU4 3CLpro bound to non-covalent inhibitor 2A | | Descriptor: | 3C-like proteinase, ACETATE ION, FORMIC ACID, ... | | Authors: | St John, S.E, Mesecar, A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
4YOG
 
 | | HKU4-3CLpro bound to non-covalent inhibitor 3B | | Descriptor: | 3C-like proteinase, ACETATE ION, N-[4-(acetylamino)phenyl]-2-(1H-benzotriazol-1-yl)-N-[(1R)-2-(tert-butylamino)-2-oxo-1-(thiophen-3-yl)ethyl]acetamide | | Authors: | St John, S.E, Mesecar, A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
4YOI
 
 | | Structure of HKU4 3CLpro bound to non-covalent inhibitor 1A | | Descriptor: | 3C-like proteinase, ACETATE ION, FORMIC ACID, ... | | Authors: | St John, S.E, Mesecar, A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
4OW0
 
 | |
4OVZ
 
 | |
3F4C
 
 | | Crystal structure of organophosphorus hydrolase from Geobacillus stearothermophilus strain 10, with glycerol bound | | Descriptor: | COBALT (II) ION, GLYCEROL, Organophosphorus hydrolase | | Authors: | Hawwa, R, Aikens, J, Turner, R.J, Santarsiero, B, Mesecar, A. | | Deposit date: | 2008-10-31 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus.
Arch.Biochem.Biophys., 488, 2009
|
|
