2IFB
 
 | |
2HH7
 
 | |
4XT8
 
 | | Crystal structure of Rv2671 from Mycobacterium tuberculosis in complex with NADP+ and trimetrexate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
4XT7
 
 | |
4XT5
 
 | |
4XT6
 
 | | Crystal structure of Rv2671 from Mycobacterium tuberculosis in complex with the tetrahydropteridine ring of tetrahydrofolate (THF) | | Descriptor: | (6S)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Rv2671 | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
4XT4
 
 | | Crystal structure of Rv2671 from Mycobacteirum tuberculosis in complex with dihydropteridine ring of dihydropteroic acid | | Descriptor: | 2-amino-6-methyl-7,8-dihydropteridin-4(3H)-one, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
3F4B
 
 | |
4R3O
 
 | | Human Constitutive 20S Proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Sacchettini, J.C, Harshbarger, W.H. | | Deposit date: | 2014-08-16 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
4R67
 
 | | Human constitutive 20S proteasome in complex with carfilzomib | | Descriptor: | N-{(2S)-2-[(morpholin-4-ylacetyl)amino]-4-phenylbutanoyl}-L-leucyl-N-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-L-phenylalaninamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Sacchettini, J.C, Harshbarger, W.H. | | Deposit date: | 2014-08-22 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
3OKR
 
 | |
1IFB
 
 | |
1M1M
 
 | |
1MAS
 
 | | PURINE NUCLEOSIDE HYDROLASE | | Descriptor: | INOSINE-URIDINE NUCLEOSIDE N-RIBOHYDROLASE, POTASSIUM ION | | Authors: | Degano, M, Gopaul, D.N, Scapin, G, Schramm, V.L, Sacchettini, J.C. | | Deposit date: | 1995-12-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of the inosine-uridine nucleoside N-ribohydrolase from Crithidia fasciculata.
Biochemistry, 35, 1996
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
8GKF
 
 | |
7RB1
 
 | | Isocitrate Lyase-1 from Mycobacterium tuberculosis covalently modified by 5-descarboxy-5-nitro-D-isocitric acid | | Descriptor: | (3E)-3-(hydroxyimino)propanoic acid, GLYCEROL, GLYOXYLIC ACID, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T, Sacchettini, J.C. | | Deposit date: | 2021-07-05 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Based Inactivation of Mycobacterium tuberculosis Isocitrate Lyase 1 by (2 R ,3 S )-2-Hydroxy-3-(nitromethyl)succinic acid.
J.Am.Chem.Soc., 143, 2021
|
|
6UX6
 
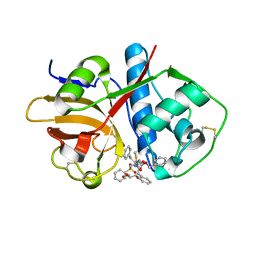 | | Cruzain covalently bound by a vinylsulfone compound | | Descriptor: | Cruzipain, GLYCEROL, Nalpha-[(benzyloxy)carbonyl]-N-[(2S)-1-phenyl-4-(phenylsulfonyl)butan-2-yl]-L-phenylalaninamide | | Authors: | Zhai, X, Tang, S, Chenna, B.C, Meek, T.D, Sacchettini, J.C. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Cruzain covalently bound by a vinylsulfone compound
To Be Published
|
|
3LF0
 
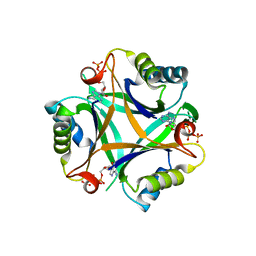 | | Crystal structure of the ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory protein P-II | | Authors: | Shetty, N.D, Palaninathan, S.K, Reddy, M.C.M, Owen, J.L, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-15 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein.
Protein Sci., 19, 2010
|
|
8W1U
 
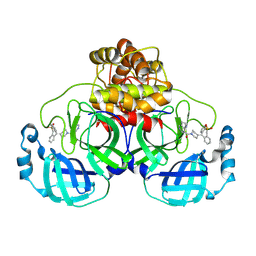 | | SARS-CoV-2 Main protease bound to non-covalent lead molecule NZ-804 | | Descriptor: | 11-[1-(1H-pyrrolo[3,2-c]pyridine-7-carbonyl)piperidin-4-ylidene]-6,11-dihydro-5H-5lambda~6~-dibenzo[b,e]thiepine-5,5-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
8W1T
 
 | | SARS-CoV-2 Main protease bound to a non-covalent non-peptidic HTS hit | | Descriptor: | (5-bromopyridin-3-yl){4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, (5-bromopyridin-3-yl){4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, 1,2-ETHANEDIOL, ... | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
4UBE
 
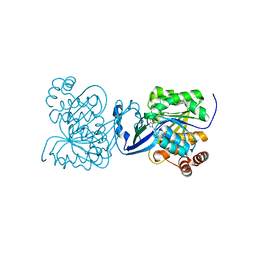 | | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C. | | Deposit date: | 2014-08-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE
TO BE PUBLISHED
|
|
3GS4
 
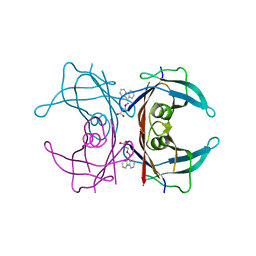 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid (inhibitor 15) | | Descriptor: | 3-[(9H-fluoren-9-ylideneamino)oxy]propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis.
Plos One, 4, 2009
|
|
3GS0
 
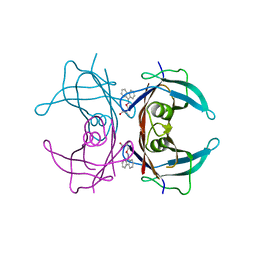 | | Human transthyretin (TTR) complexed with (S)-3-(9H-fluoren-9-ylideneaminooxy)-2-methylpropanoic acid (inhibitor 16) | | Descriptor: | (2S)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methylpropanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis.
Plos One, 4, 2009
|
|
3GLZ
 
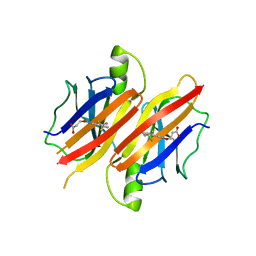 | | Human Transthyretin (TTR) complexed with(E)-3-(2-(trifluoromethyl)benzylideneaminooxy)propanoic acid (inhibitor 11) | | Descriptor: | 3-[({(1E)-[2-(trifluoromethyl)phenyl]methylidene}amino)oxy]propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis
Plos One, 4, 2009
|
|
