1NNT
 
 | |
3GS7
 
 | | Human transthyretin (TTR) complexed with (E)-3-(2-methoxybenzylideneaminooxy)propanoic acid (inhibitor 13) | | Descriptor: | 3-({[(1Z)-(2-methoxyphenyl)methylidene]amino}oxy)propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis.
Plos One, 4, 2009
|
|
3U39
 
 | | Crystal Structure of the apo Bacillus Stearothermophilus phosphofructokinase | | Descriptor: | 6-phosphofructokinase, CALCIUM ION | | Authors: | Mosser, R, Reddy, M.C.M, Bruning, J.B, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7921 Å) | | Cite: | Structure of the apo form of Bacillus stearothermophilus phosphofructokinase.
Biochemistry, 51, 2012
|
|
3ONR
 
 | | Crystal structure of the calcium chelating immunodominant antigen, calcium dodecin (Rv0379),from Mycobacterium tuberculosis with a novel calcium-binding site | | Descriptor: | CALCIUM ION, FORMIC ACID, PLATINUM (II) ION, ... | | Authors: | Arulandu, A, Sacchettini, J.C. | | Deposit date: | 2010-08-30 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of calcium dodecin (Rv0379), from Mycobacterium tuberculosis with a unique calcium-binding site.
Protein Sci., 20, 2011
|
|
1KPH
 
 | | Crystal Structure of mycolic acid cyclopropane synthase CmaA1 complexed with SAH and DDDMAB | | Descriptor: | CARBONATE ION, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1, DIDECYL-DIMETHYL-AMMONIUM, ... | | Authors: | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-30 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
1KPG
 
 | | Crystal Structure of mycolic acid cyclopropane synthase CmaA1 complexed with SAH and CTAB | | Descriptor: | CARBONATE ION, CETYL-TRIMETHYL-AMMONIUM, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1, ... | | Authors: | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-30 | | Release date: | 2002-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
3PZK
 
 | |
3Q7U
 
 | | Structure of Mtb 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (IspD) complexed with CTP | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Ioerger, T.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-01-05 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (IspD): a candidate antitubercular drug target
Proteins, 2011
|
|
3QJA
 
 | |
4EDH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg.
To be Published
|
|
4KBM
 
 | |
4KBJ
 
 | |
3RKJ
 
 | | Crystal Structure of New Delhi Metallo-Beta-Lactamase-1 from Klebsiella pnueumoniae | | Descriptor: | Beta-lactamase NDM-1, GLYCEROL, SULFATE ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Binkowski, T.A, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3RKK
 
 | | Crystal Structure of New Delhi Metallo-Beta-Lactamase-1 from Klebsiella pneumoniae | | Descriptor: | ACETIC ACID, Beta-lactamase NDM-1, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Binkowski, T.A, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
1N8W
 
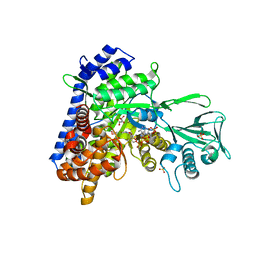 | | Biochemical and Structural Studies of Malate Synthase from Mycobacterium tuberculosis | | Descriptor: | COENZYME A, D-MALATE, GLYOXYLIC ACID, ... | | Authors: | Smith, C.V, Huang, C.C, Miczak, A, Russell, D.G, Sacchettini, J.C, Honer zu Bentrup, K, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-11-21 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis
J.Biol.Chem., 278, 2003
|
|
4GMD
 
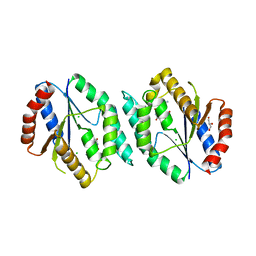 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with AZT Monophosphate
To be Published
|
|
3T78
 
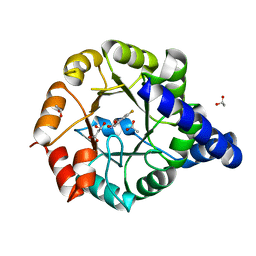 | | Crystal Structure of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS) in Complex with 5-Fluoroanthranilate | | Descriptor: | 2-amino-5-fluorobenzoic acid, ACETATE ION, Indole-3-glycerol phosphate synthase, ... | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights and Inhibition of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS): an Essential Enzyme for Tryptophan Biosynthesis
To be Published
|
|
3T44
 
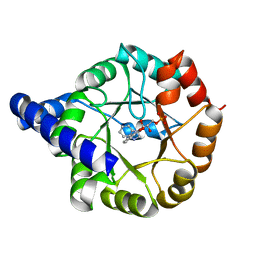 | | Crystal structure of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS) in complex with indole glycerol phosphate (IGP) amd anthranilate | | Descriptor: | 2-AMINOBENZOIC ACID, INDOLE-3-GLYCEROL PHOSPHATE, Indole-3-glycerol phosphate synthase | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-07-25 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights and Inhibition of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS): an Essential Enzyme for Tryptophan Biosynthesis
To be Published
|
|
3T55
 
 | | Crystal structure of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS) in complex with Phenoxymethyl Benzoic Acid (PMBA) | | Descriptor: | 3-(phenoxymethyl)benzoic acid, Indole-3-glycerol phosphate synthase, MALONATE ION, ... | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-07-26 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Insights and Inhibition of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS): an Essential Enzyme for Tryptophan Biosynthesis
To be Published
|
|
3T40
 
 | | Crystal Structure of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS) complex with N-2-Carboxyphenyl Glycine (CPG) | | Descriptor: | 2-[(carboxymethyl)amino]benzoic acid, Indole-3-glycerol phosphate synthase, SULFATE ION | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-07-25 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural Insights and Inhibition of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS): an Essential Enzyme for Tryptophan Biosynthesis
To be Published
|
|
1F2V
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PRECORRIN-8X METHYLMUTASE OF AEROBIC VITAMIN B12 SYNTHESIS | | Descriptor: | PRECORRIN-8X METHYLMUTASE | | Authors: | Shipman, L.W, Li, D, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2000-05-29 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of precorrin-8x methyl mutase.
Structure, 9, 2001
|
|
1FPS
 
 | |
1R8J
 
 | |
1LD9
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN H-2LD PEPTIDE COMPLEX EXPLAINS THE UNIQUE INTERACTION OF LD WITH BETA2M AND PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2LD HEAVY CHAIN, NANO-PEPTIDE | | Authors: | Balendiran, G.K, Solheim, J.C, Young, A.C.M, Hansen, T.H, Nathenson, S.G, Sacchettini, J.C. | | Deposit date: | 1997-04-24 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of an H-2Ld-peptide complex explains the unique interaction of Ld with beta-2 microglobulin and peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1XJT
 
 | |
