5UF6
 
 | | The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, capsid protein VP1 | | Authors: | Xie, Q, Spear, J.M, Noble, A.J, Sousa, D.R, Meyer, N.L, Davulcu, O, Zhang, F, Linhardt, R.J, Stagg, S.M, Chapman, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The 2.8 angstrom Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide.
Mol Ther Methods Clin Dev, 5, 2017
|
|
3BEJ
 
 | | Structure of human FXR in complex with MFA-1 and co-activator peptide | | Descriptor: | (8alpha,10alpha,13alpha,17beta)-17-[(4-hydroxyphenyl)carbonyl]androsta-3,5-diene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Soisson, S.M, Parthasarathy, G, Becker, J.W. | | Deposit date: | 2007-11-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4JSW
 
 | | Human carbonic anhydrase II H94C | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Martin, D.P, Hann, Z.S, Cohen, S.M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metalloprotein-Inhibitor Binding: Human Carbonic Anhydrase II as a Model for Probing Metal-Ligand Interactions in a Metalloprotein Active Site.
Inorg.Chem., 52, 2013
|
|
3PME
 
 | | Crystal structure of the receptor binding domain of botulinum neurotoxin C/D mosaic serotype | | Descriptor: | GLYCEROL, SULFATE ION, Type C neurotoxin | | Authors: | Zhang, Y, Buchko, G.W, Qin, L, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-11-16 | | Release date: | 2010-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the receptor binding domain of the botulinum C-D mosaic neurotoxin reveals potential roles of lysines 1118 and 1136 in membrane interactions.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
6H37
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenyl)-4-hydroxy-1-piperidine-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-(4-oxidanyl-4-phenyl-piperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
3PUF
 
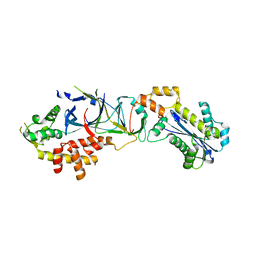 | | Crystal structure of human RNase H2 complex | | Descriptor: | Ribonuclease H2 subunit A, Ribonuclease H2 subunit B, Ribonuclease H2 subunit C | | Authors: | Figiel, M, Chon, H, Cerritelli, S.M, Cybulska, M, Crouch, R.J, Nowotny, M. | | Deposit date: | 2010-12-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects.
J.Biol.Chem., 286, 2011
|
|
3B6R
 
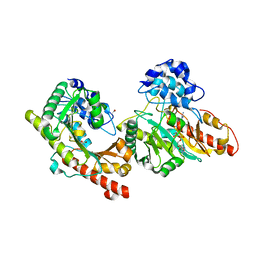 | | Crystal structure of Human Brain-type Creatine Kinase | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Creatine kinase B-type, ... | | Authors: | Bong, S.M, Moon, J.H, Hwang, K.Y. | | Deposit date: | 2007-10-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3- -creatine transition-state analogue complex
Febs Lett., 582, 2008
|
|
4JS6
 
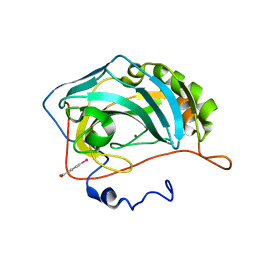 | | Crystal structure of inhibitor-free hCAII H94D | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Martin, D.P, Hann, Z.S, Cohen, S.M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metalloprotein-Inhibitor Binding: Human Carbonic Anhydrase II as a Model for Probing Metal-Ligand Interactions in a Metalloprotein Active Site.
Inorg.Chem., 52, 2013
|
|
5WNX
 
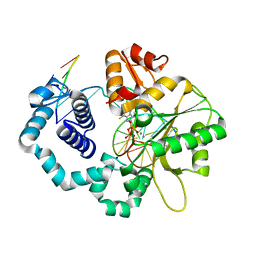 | | DNA polymerase beta substrate complex with incoming 6-TdGTP | | Descriptor: | 2-amino-9-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-1,9-dihydro-6H-purine-6-thione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
4FJ4
 
 | | Crystal structure of the protein Q9HRE7 complexed with mercury from Halobacterium salinarium at the resolution 2.1A, Northeast Structural Genomics Consortium target HsR50 | | Descriptor: | ETHYL MERCURY ION, SODIUM ION, Uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Maglaqui, M, Owens, L.A, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
3DR5
 
 | | Crystal structure of the Q8NRD3_CORGL protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium target CgR117. | | Descriptor: | Putative O-Methyltransferase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Wang, D, Ciccosanti, C, Foote, E.L, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-10 | | Release date: | 2008-09-09 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Q8NRD3_CORGL protein from Corynebacterium glutamicum.
To be Published
|
|
3DT7
 
 | |
3RGB
 
 | |
5TZL
 
 | | Structure of transthyretin in complex with the kinetic stabilizer 201 | | Descriptor: | 4-(7-chloro-1,3-benzoxazol-2-yl)-2,6-diiodophenol, Transthyretin | | Authors: | Connelly, S, Mortenson, D.E, Choi, S, Wilson, I.A, Powers, E.T, Kelly, J.W, Johnson, S.M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Semi-quantitative models for identifying potent and selective transthyretin amyloidogenesis inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TZY
 
 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8BLO
 
 | | Human Urea Transporter UT-A (N-Terminal Domain Model) | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Urea transporter 2, di-heneicosanoyl phosphatidyl choline | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Scacioc, A, Wang, D, McKinley, G, Fernandez-Cid, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Burgess-Brown, N.A, van Putte, W, Duerr, K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
8BLP
 
 | | Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14 | | Descriptor: | 10-(4-ethylphenyl)sulfonyl-~{N}-(thiophen-2-ylmethyl)-5-thia-1,8,11,12-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,7,9,11-pentaen-7-amine, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Chi, G, Dietz, L, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, Scacioc, A, McKinley, G, Arrowsmith, C.H, Edwards, A, Bountra, C, Fernandez-Cid, A, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
4EGU
 
 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
6I84
 
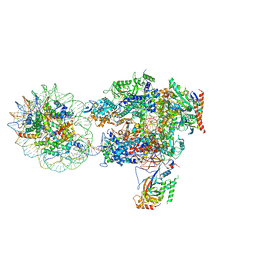 | | Structure of transcribing RNA polymerase II-nucleosome complex | | Descriptor: | DNA (158-MER), DNA (170-MER), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Farnung, L, Vos, S.M, Cramer, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of transcribing RNA polymerase II-nucleosome complex.
Nat Commun, 9, 2018
|
|
4E4A
 
 | |
5WO0
 
 | | DNA polymerase beta substrate complex with incoming 5-FodUTP | | Descriptor: | 2'-deoxy-5-formyluridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
6KL5
 
 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
3R3R
 
 | | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica | | Descriptor: | ZINC ION, ferripyochelin binding protein | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica
TO BE PUBLISHED
|
|
3RRX
 
 | | Crystal Structure of Q683A mutant of Exo-1,3/1,4-beta-glucanase (ExoP) from Pseudoalteromonas sp. BB1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2011-05-01 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 279, 2012
|
|
4ELL
 
 | |
