1G1E
 
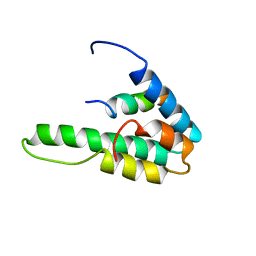 | | NMR STRUCTURE OF THE HUMAN MAD1 TRANSREPRESSION DOMAIN SID IN COMPLEX WITH MAMMALIAN SIN3A PAH2 DOMAIN | | Descriptor: | MAD1 PROTEIN, SIN3A | | Authors: | Brubaker, K, Cowley, S.M, Huang, K, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
7KQL
 
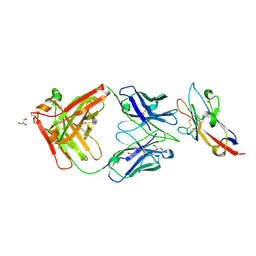 | | Anti-Tim3 antibody Fab complex | | Descriptor: | GLYCEROL, Hepatitis A virus cellular receptor 2, Tim3.18 Fab heavy chain, ... | | Authors: | Deng, X.A, West, S.M, Strop, P. | | Deposit date: | 2020-11-16 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Tim-3 mediates T cell trogocytosis to limit antitumor immunity.
J.Clin.Invest., 132, 2022
|
|
3U23
 
 | | Atomic resolution crystal structure of the 2nd SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide from human RIN3 | | Descriptor: | 1,2-ETHANEDIOL, CD2-associated protein, Ras and Rab interactor 3 | | Authors: | Simister, P.C, Rouka, E, Janning, M, Muniz, J.R.C, Kirsch, K.H, Knapp, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Krojer, T, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
4MAC
 
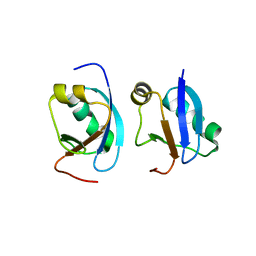 | | Crystal structure of CIDE-N domain of FSP27 | | Descriptor: | Cell death activator CIDE-3 | | Authors: | Park, H.H, Lee, S.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for homo-dimerization of the CIDE domain revealed by the crystal structure of the CIDE-N domain of FSP27
Biochem.Biophys.Res.Commun., 439, 2013
|
|
7KZE
 
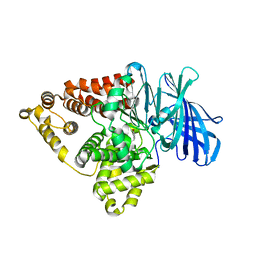 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, Leukotriene A-4 hydrolase, TRIETHYLENE GLYCOL, ... | | Authors: | Lee, K.H, Shim, Y, Paige, M, Noble, S.M. | | Deposit date: | 2020-12-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
2W2T
 
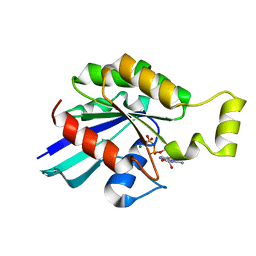 | | Rac2 (G12V) in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 2 | | Authors: | Opaleye, O, Bunney, T.D, Roe, S.M, Pearl, L.H. | | Deposit date: | 2008-11-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into Formation of an Active Signaling Complex between Rac and Phospholipase C Gamma 2.
Mol.Cell, 34, 2009
|
|
2W2V
 
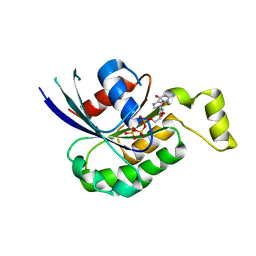 | | Rac2 (G12V) in complex with GTPgS | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 2 | | Authors: | Opaleye, O, Bunney, T.D, Roe, S.M, Pearl, L.H. | | Deposit date: | 2008-11-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Formation of an Active Signaling Complex between Rac and Phospholipase C Gamma 2.
Mol.Cell, 34, 2009
|
|
4LU3
 
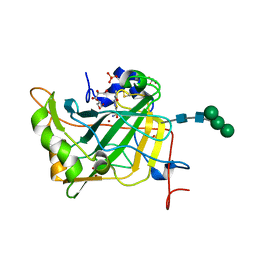 | | The crystal structure of the human carbonic anhydrase XIV | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 14, GLYCEROL, ... | | Authors: | Alterio, V, De Simone, G, Monti, S.M. | | Deposit date: | 2013-07-24 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural comparison between membrane-associated human carbonic anhydrases provides insights into drug design of selective inhibitors.
Biopolymers, 101, 2014
|
|
2WEQ
 
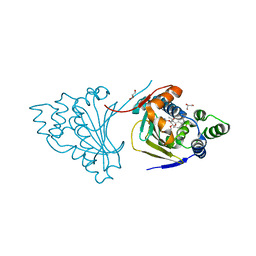 | |
2WEP
 
 | |
5G21
 
 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 26). | | Descriptor: | ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G1Z
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 1) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
4M0L
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Arkhipova, V.I, Gabdulkhakov, A.G, Nikulin, A.D, Lazopulo, A.M, Lazopulo, S.M, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational transitions in the gamma subunit of the archaeal translation initiation factor 2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M33
 
 | | Crystal structure of gated-pore mutant H141D of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
2WHJ
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, GLYCEROL, ... | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
1HY7
 
 | | A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 | | Descriptor: | CALCIUM ION, R-2-{[4'-METHOXY-(1,1'-BIPHENYL)-4-YL]-SULFONYL}-AMINO-6-METHOXY-HEX-4-YNOIC ACID, STROMELYSIN-1, ... | | Authors: | Natchus, M.G, Bookland, R.G, Laufersweiler, M.J, Pikul, S, Almstead, N.G, De, B, Janusz, M.J, Hsieh, L.C, Gu, F, Pokross, M.E, Patel, V.S, Garver, S.M, Peng, S.X, Branch, T.M, King, S.L, Baker, T.R, Foltz, D.J, Mieling, G.E. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of new carboxylic acid-based MMP inhibitors derived from functionalized propargylglycines.
J.Med.Chem., 44, 2001
|
|
7L7Q
 
 | | Ctf3c with Ulp2-KIM | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2020-12-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ctf3/CENP-I provides a docking site for the desumoylase Ulp2 at the kinetochore.
J.Cell Biol., 220, 2021
|
|
3UAN
 
 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4LPA
 
 | | Crystal structure of a Cdc6 phosphopeptide in complex with Cks1 | | Descriptor: | Cyclin-dependent kinases regulatory subunit | | Authors: | McGrath, D.A, Balog, E.R.M, Koivomagi, M, Lucena, R, Mai, M.V, Hirschi, A, Kellogg, D.R, Loog, M, Rubin, S.M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cks confers specificity to phosphorylation-dependent CDK signaling pathways.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2WMO
 
 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
8R36
 
 | | Crystal structure of the Gluk1 ligand-binding domain in complex with kainate and BPAM538 at 1.90 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Bay, Y, Frantsen, S.M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GluK1 ligand-binding domain with kainate and the full-spanning positive allosteric modulator BPAM538.
J.Struct.Biol., 216, 2024
|
|
4LTN
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN, NADH complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, ... | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
5H46
 
 | | Mycobacterium smegmatis Dps1 mutant - F47E | | Descriptor: | DNA protection during starvation protein, FE (II) ION | | Authors: | Williams, S.M, Chandran, A.V, Vijayan, M, Chatterji, D. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Mutation Directs the Structural Switch of DNA Binding Proteins under Starvation to a Ferritin-like Protein Cage.
Structure, 25, 2017
|
|
7FQT
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRP
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with XST00000245b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(2-methyl-1,3-thiazol-4-yl)thiophene-2-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
