1AC3
 
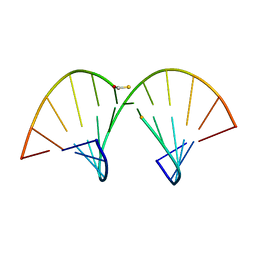 | | SOLUTION STRUCTURE OF AN RNA-DNA HYBRID DUPLEX CONTAINING A 3'-THIOFORMACETAL LINKER AND AN RNA A-TRACT, NMR, 8 STRUCTURES | | Descriptor: | ANTISENSE HYBRID DUPLEX, DNA (5'-D(*(TCP)P*TP*GP*CP*GP*C)-3') | | Authors: | Cross, C.W, Rice, J.S, Gao, X. | | Deposit date: | 1997-02-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA x DNA hybrid duplex containing a 3'-thioformacetal linker and an RNA A-tract.
Biochemistry, 36, 1997
|
|
7MI8
 
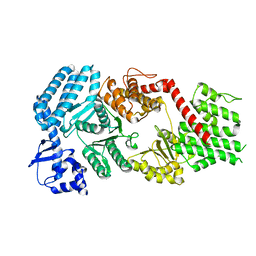 | | Signal subtracted reconstruction of AAA5 and AAA6 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 5 | | Descriptor: | Fusion protein of Dynein and Endolysin | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI1
 
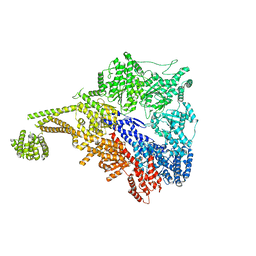 | |
7MI3
 
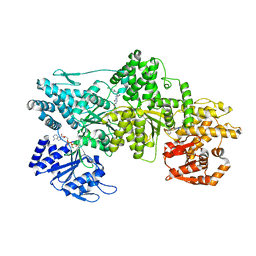 | | Signal subtracted reconstruction of AAA2, AAA3, and AAA4 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 4 | | Descriptor: | (8S)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile, ADENOSINE-5'-TRIPHOSPHATE, Fusion protein of Dynein and Endolysin, ... | | Authors: | Santarossa, C.C, Coudray, N, Urnavicius, L, Ekiert, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
7MI6
 
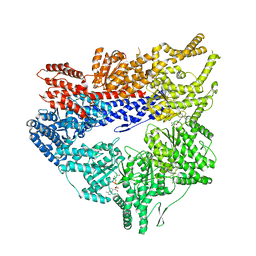 | | Yeast dynein motor domain in the presence of a pyrazolo-pyrimidinone-based compound, Model 1 | | Descriptor: | (8S)-6-(3-bromophenoxy)-2-[1-(4-chlorophenyl)cyclopropyl]-7-hydroxypyrazolo[1,5-a]pyrimidine-3-carbonitrile, ADENOSINE-5'-TRIPHOSPHATE, Fusion protein of Dynein and Endolysin, ... | | Authors: | Santarossa, C.C, Urnavicius, L, Coudray, N, Ekeirt, D.C, Bhabha, G, Kapoor, T.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Targeting allostery in the Dynein motor domain with small molecule inhibitors.
Cell Chem Biol, 28, 2021
|
|
8RCW
 
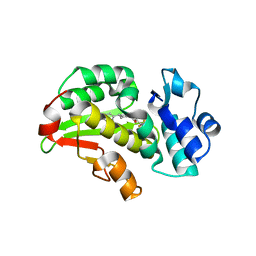 | | Crystal structure of the Mycobacterium tuberculosis regulator VirS (N-terminal fragment 4-208) in complex with the lead compound SMARt751 | | Descriptor: | 4,4,4-tris(fluoranyl)-1-[4-(4-fluorophenyl)piperidin-1-yl]butan-1-one, HTH-type transcriptional regulator VirS | | Authors: | Grosse, C, Sigoillot, M, Megalizzi, V, Tanina, A, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2023-12-07 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis VirS regulator reveals its interaction with the lead compound SMARt751.
J.Struct.Biol., 216, 2024
|
|
3UFE
 
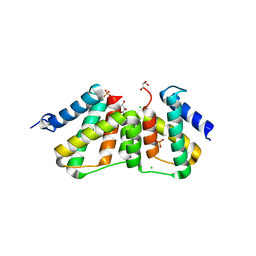 | | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grosse, C, Himmel, S, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2011-11-01 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution
To be Published
|
|
1L5P
 
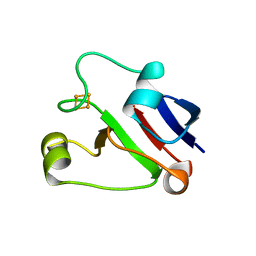 | | Crystal Structure of Trichomonas vaginalis Ferredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, ferredoxin | | Authors: | Crossnoe, C.R, Germanas, J.P, Le Magueres, P, Mustata, G, Krause, K.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-08-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Trichomonas vaginalis ferredoxin provides insight into metronidazole activation.
J.Mol.Biol., 318, 2002
|
|
1VRU
 
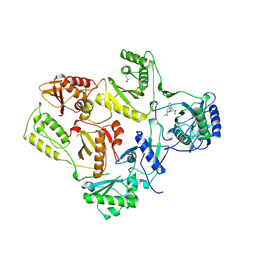 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
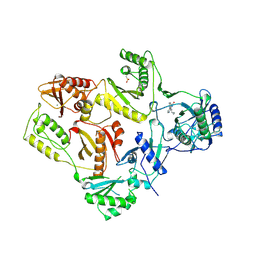 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1RT2
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1RT1
 
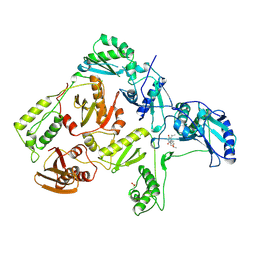 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH MKC-442 | | Descriptor: | 6-BENZYL-1-ETHOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Willcox, B, Jones, Y, Ross, C, Stammers, D, Stuart, D. | | Deposit date: | 1996-03-16 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors.
J.Med.Chem., 39, 1996
|
|
1REV
 
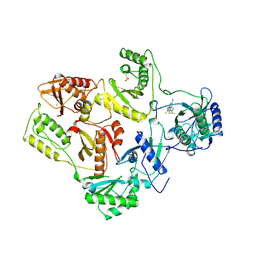 | | HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-09-17 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design.
Structure, 3, 1995
|
|
1RTI
 
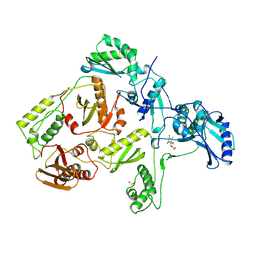 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1RTH
 
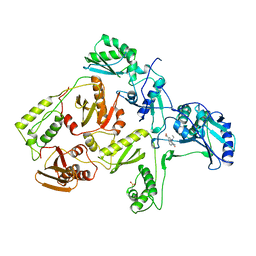 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1RTJ
 
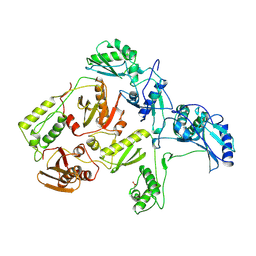 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
1CX5
 
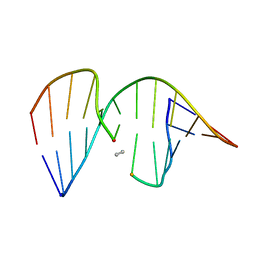 | | ANTISENSE DNA/RNA HYBRID CONTAINING MODIFIED BACKBONE | | Descriptor: | 5'-D(*CP*GP*CP*GP*TP*T*(MMT)P*TP*GP*CP*GP*C), 5'-R(*GP*CP*GP*CP*AP*AP*AP*AP*CP*GP*CP*G) | | Authors: | Yang, X, Han, X, Cross, C, Sanghvi, Y, Gao, X. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an antisense DNA.RNA hybrid duplex containing a 3'-CH(2)N(CH(3))-O-5' or an MMI backbone linker.
Biochemistry, 38, 1999
|
|
1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
1AZG
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, MINIMIZED AVERAGE (PROBMAP) STRUCTURE | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
3OEO
 
 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
2XI8
 
 | | High resolution structure of native CylR2 | | Descriptor: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-06-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XIU
 
 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XJ3
 
 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
1S16
 
 | | Crystal Structure of E. coli Topoisomerase IV ParE 43kDa subunit complexed with ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Wei, Y, Gross, C.H. | | Deposit date: | 2004-01-05 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase.
Antimicrob.Agents Chemother., 48, 2004
|
|
1S14
 
 | | Crystal structure of Escherichia coli Topoisomerase IV ParE 24kDa subunit | | Descriptor: | NOVOBIOCIN, Topoisomerase IV subunit B | | Authors: | Wei, Y, Gross, C.H. | | Deposit date: | 2004-01-05 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase.
Antimicrob.Agents Chemother., 48, 2004
|
|
