5OPK
 
 | | Crystal structure of D52N/R367Q cN-II mutant bound to dATP and free phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPP
 
 | | Crystal structure of S408R cN-II mutant | | Descriptor: | BROMIDE ION, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPN
 
 | | Crystal structure of R39Q cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPO
 
 | | Crystal structure of R238G cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, MAGNESIUM ION | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
4WDQ
 
 | | Crystal structure of haloalkane dehalogenase LinB32 mutant (L177W) from Sphingobium japonicum UT26 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, MAGNESIUM ION | | Authors: | Degtjarik, O, Rezacova, P, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WCV
 
 | | Haloalkane dehalogenase DhaA mutant from Rhodococcus rhodochrous (T148L+G171Q+A172V+C176G) | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Holubeva, T, Prudnikova, T, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2014-09-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Balancing the stability-activity trade-off by fine-tuning dehalogenase access tunnels
Chemcatchem, 2015
|
|
4WDR
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
7QHL
 
 | | Crystal structure of Cyclin-dependent kinase 2/cyclin A in complex with 3,5,7-Substituted pyrazolo[4,3-d]pyrimidine inhibitor 24 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-amino-1-ethyl)thio-3-cyclobutyl-7-[4-(pyrazol-1-yl)benzyl]amino-1(2)H-pyrazolo[4,3-d]pyrimidine, Cyclin-A2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d ]Pyrimidine Inhibitors of Cyclin-Dependent Kinases and Cyclin K Degraders.
J.Med.Chem., 65, 2022
|
|
3FV3
 
 | | Secreted aspartic protease 1 from Candida parapsilosis in complex with pepstatin A | | Descriptor: | GLYCEROL, SULFATE ION, Sapp1p-secreted aspartic protease 1, ... | | Authors: | Dostal, J, Brynda, J, Hruskova-Heidingsfeldova, O, Sieglova, I, Pichova, I, Rezacova, P. | | Deposit date: | 2009-01-15 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the secreted aspartic protease 1 from Candida parapsilosis in complex with pepstatin A
J.Struct.Biol., 167, 2009
|
|
1NH0
 
 | | 1.03 A structure of HIV-1 protease: inhibitor binding inside and outside the active site | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEASE RETROPEPSIN, SULFATE ION, ... | | Authors: | Brynda, J, Rezacova, P, Fabry, M, Horejsi, M, Hradilek, M, Soucek, M, Konvalinka, J, Sedlacek, J. | | Deposit date: | 2002-12-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A Phenylnorstatine Inhibitor Binding to HIV-1 Protease: Geometry,
Protonation, and Subsite-Pocket Interactions Analyzed at Atomic Resolution
J.Med.Chem., 47, 2004
|
|
1N4X
 
 | | Structure of scFv 1696 at acidic pH | | Descriptor: | CHLORIDE ION, immunoglobulin heavy chain variable region, immunoglobulin kappa chain variable region | | Authors: | Lescar, J, Brynda, J, Fabry, M, Horejsi, M, Rezacova, P, Sedlacek, J, Bentley, G.A. | | Deposit date: | 2002-11-02 | | Release date: | 2003-06-10 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a single-chain Fv fragment of an antibody that inhibits the HIV-1 and HIV-2 proteases.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7ACK
 
 | | CDK2/cyclin A2 in complex with an imidazo[1,2-c]pyrimidin-5-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclohexyl-6~{H}-imidazo[1,2-c]pyrimidin-5-one, Cyclin-A2, ... | | Authors: | Skerlova, J, Pachl, P, Rezacova, P. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Imidazo[1,2-c]pyrimidin-5(6H)-one inhibitors of CDK2: Synthesis, kinase inhibition and co-crystal structure.
Eur.J.Med.Chem., 216, 2021
|
|
3KRV
 
 | | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rakonjac, N, Rezacova, P, Borek, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure Of Potential Metal-Dependent Hydrolase With Cyclase Activity
To be Published
|
|
5K7Y
 
 | | Crystal structure of enzyme in purine metabolism | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Skerlova, J, Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2016-05-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interface modulation causes misregulation of purine 5 -nucleotidase in relapsed leukemia.
Bmc Biol., 14, 2016
|
|
3I8T
 
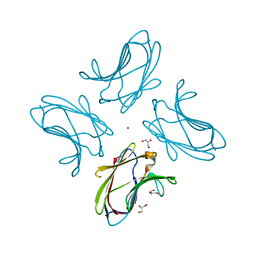 | | N-terminal CRD1 domain of mouse Galectin-4 in complex with lactose | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Galectin-4, ... | | Authors: | Brynda, J, Krejcirikova, V, Rezacova, P. | | Deposit date: | 2009-07-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the mouse galectin-4 N-terminal carbohydrate-recognition domain reveals the mechanism of oligosaccharide recognition
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5L4Z
 
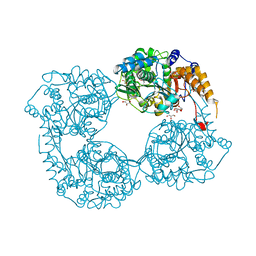 | |
5L50
 
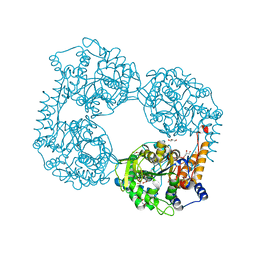 | |
5LKA
 
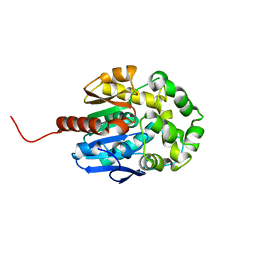 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
7BHY
 
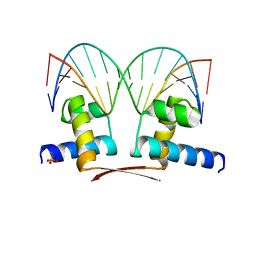 | | DNA-binding domain of DeoR in complex with the DNA operator | | Descriptor: | DNA operator - strand 1, DNA operator - strand 2, Deoxyribonucleoside regulator, ... | | Authors: | Novakova, M, Rezacova, P, Skerlova, J, Brynda, J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into DNA recognition by bacterial transcriptional regulators of the SorC/DeoR family.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3GGU
 
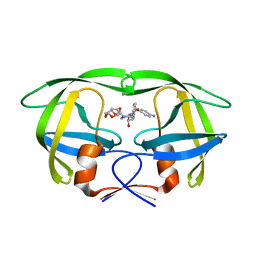 | | HIV PR drug resistant patient's variant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Saskova, K.G, Brynda, J, Rezacova, P. | | Deposit date: | 2009-03-02 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular characterization of clinical isolates of human immunodeficiency virus resistant to the protease inhibitor darunavir.
J.Virol., 83, 2009
|
|
6ZTK
 
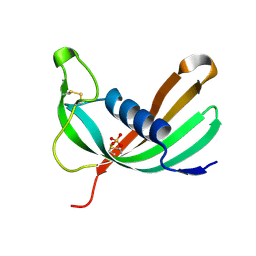 | | Crystal structure of Mialostatin, a gut cystatin from the hard tick Ixodes ricinus | | Descriptor: | FRAGMENT OF TRITON X-100, Mialostatin, SULFATE ION | | Authors: | Busa, M, Rezacova, P, Mares, M. | | Deposit date: | 2020-07-20 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mialostatin, a Novel Midgut Cystatin from Ixodes ricinus Ticks: Crystal Structure and Regulation of Host Blood Digestion.
Int J Mol Sci, 22, 2021
|
|
7B7S
 
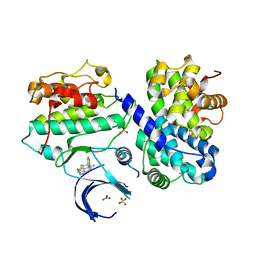 | | CDK2/cyclin A2 in complex with 3H-pyrazolo[4,3-f]quinoline-based derivative HSD1368 | | Descriptor: | 7-(3-(trifluoromethyl)-1H-pyrazol-4yl)-3,8,10,11-tetrahydropyrazolo[4,3-f]thiopyrano[3,4-c]quinoline 9-oxide, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3 H -Pyrazolo[4,3- f ]quinoline-Based Kinase Inhibitors Inhibit the Proliferation of Acute Myeloid Leukemia Cells In Vivo.
J.Med.Chem., 64, 2021
|
|
7ZSO
 
 | | human purine nucleoside phosphorylase in complex with JS-554 | | Descriptor: | GLYCEROL, Purine nucleoside phosphorylase, SODIUM ION, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSL
 
 | | human purine nucleoside phosphorylase in complex with JS-196 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Purine nucleoside phosphorylase, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSM
 
 | | human purine nucleoside phosphorylase in complex with JS-375 | | Descriptor: | CHLORIDE ION, Purine nucleoside phosphorylase, SULFATE ION, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
