5FOH
 
 | | Crystal structure of the catalytic domain of NcLPMO9A | | Descriptor: | COPPER (II) ION, LITHIUM ION, POLYSACCHARIDE MONOOXYGENASE, ... | | Authors: | Westereng, B, Kracun, S.K, Dimarogona, M, Mathiesen, G, Willats, W.G.T, Sandgren, M, Aachmann, F.L, Eijsink, V.G.H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of three seemingly similar lytic polysaccharide monooxygenases fromNeurospora crassasuggests different roles in plant biomass degradation.
J.Biol.Chem., 2019
|
|
6MST
 
 | | Cryo-EM structure of human AA amyloid fibril | | Descriptor: | Serum amyloid A-1 protein | | Authors: | Loerch, S, Rennegarbe, M, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
8RKS
 
 | | Structure of VPS29-VPS35 bound to the LFa motif R21 of Fam21. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35, WASH complex subunit 2A | | Authors: | Romano-Moreno, M, Astorga-Simon, E.N, Rojas, A.L, Hierro, A. | | Deposit date: | 2023-12-30 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Retromer-mediated recruitment of the WASH complex involves discrete interactions between VPS35, VPS29, and FAM21.
Protein Sci., 33, 2024
|
|
6NUL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: N137A REDUCED (150K) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
6VET
 
 | | Human insulin analog: [GluB10,HisA8,ArgA9,TyrB20]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VEQ
 
 | | Con-Ins G1 in complex with the human insulin microreceptor in turn in complex with Fv 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Con-Ins G1 B chain, Con-Ins G1a A chain, ... | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OUO
 
 | | RF2 accommodated state bound 70S complex at long incubation time | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-01 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ORL
 
 | | RF1 pre-accommodated 70S complex at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
7UYV
 
 | | Crystal structure of JAK3 kinase domain in complex with compound 25 | | Descriptor: | 6-{[(2M)-2-(2-chloro-6-fluorophenyl)-5-oxo-5H-pyrrolo[3,4-b]pyridin-4-yl]amino}-N-ethylpyridine-3-carboxamide, CHLORIDE ION, Tyrosine-protein kinase JAK3 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ZWT
 
 | | Crystal structure of DNA-binding domain of OmpR of two-component system of Acinetobacter baumannii | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Lucchini, V, Trebosc, V, Gartenmann, S, Pieren, M, Kemmer, C, Kammerer, R.A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis.
Virulence, 13, 2022
|
|
6VER
 
 | | Human insulin analog: [GluB10,TyrB20]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.047 Å) | | Cite: | Mini-Ins: A minimal, bioactive insulin analog with alternative binding modes
not published
|
|
6VES
 
 | | Human insulin analog: [GluB10,HisA8,ArgA9]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mini-Ins: A Minimal, Bioactive Insulin Analog with Alternative Binding Modes
To Be Published
|
|
5AAR
 
 | |
8PX5
 
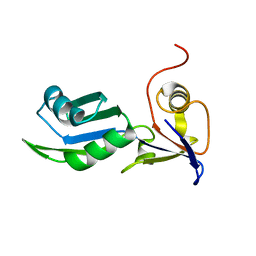 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
5AAN
 
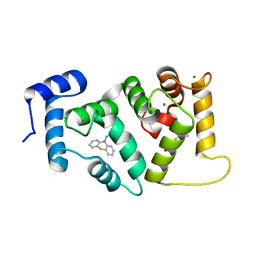 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD44 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CG5907-PA, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2015-07-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6VWI
 
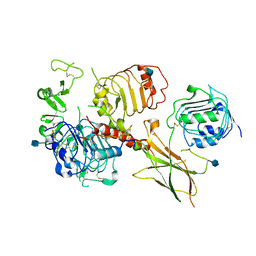 | | Head region of the closed conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor II, Leucine-zippered human type 1 insulin-like growth factor receptor ectodomain | | Authors: | Xu, Y, Kirk, N.S, Lawrence, M.C, Croll, T.I. | | Deposit date: | 2020-02-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Structure, 28, 2020
|
|
6VWH
 
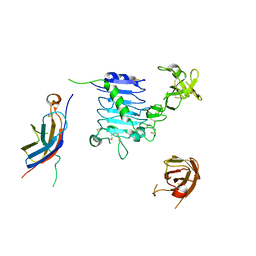 | |
8Q30
 
 | | Sulfolobus acidocaldarius AAP filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isupov, M.N, Gaines, M, Daum, B, McLaren, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | CryoEM reveals the structure of an archaeal pilus involved in twitching motility.
Nat Commun, 15, 2024
|
|
6VWG
 
 | | Head region of the open conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor II, Leucine-zippered human type 1 insulin-like growth factor receptor ectodomain | | Authors: | Xu, Y, Kirk, N.S, Lawrence, M.C, Croll, T.I. | | Deposit date: | 2020-02-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Structure, 28, 2020
|
|
7U23
 
 | |
