3K89
 
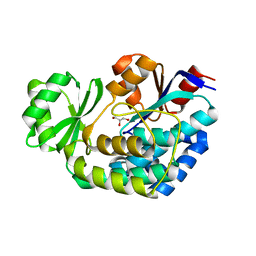 | |
1P9N
 
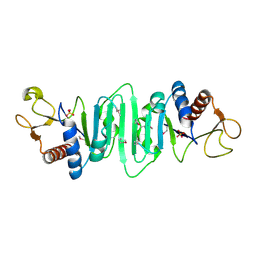 | | Crystal structure of Escherichia coli MobB. | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Rangarajan, S.E, Tocilj, A, Li, Y, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecules of Escherichia coli MobB assemble into densely packed hollow cylinders in a crystal lattice with 75% solvent content.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3V7J
 
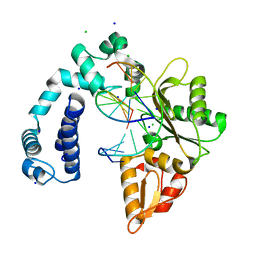 | | Co-crystal structure of Wild Type Rat polymerase beta: Enzyme-DNA binary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*AP*AP*CP*TP*CP*AP*CP*AP*TP*A)-3'), ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
3V7K
 
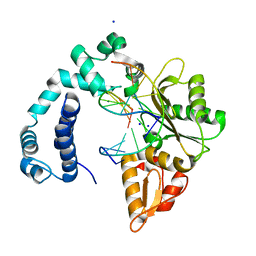 | |
3V7L
 
 | | Apo Structure of Rat DNA polymerase beta K72E variant | | Descriptor: | CHLORIDE ION, DNA polymerase beta, SODIUM ION, ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
2I0H
 
 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5T1Z
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Ethoxytriphenylethylene and GRIP Peptide | | Descriptor: | 4,4'-[(1Z)-1-(4-ethoxyphenyl)but-1-ene-1,2-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Fanning, S.W, Rajan, S.S, Maximov, P.Y, Abderrahman, B.H, Surojeet, S, Fernandes, D.J, Fan, P, Curpan, R.F, Greene, G.L, Jordan, V.C. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Endoxifen, 4-Hydroxytamoxifen and an Estrogenic Derivative Modulate Estrogen Receptor Complex Mediated Apoptosis in Breast Cancer.
Mol. Pharmacol., 94, 2018
|
|
4EC2
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
3OEQ
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, with full length n-terminus | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3OER
 
 | | Crystal structure of trimeric frataxin from the yeast saccharomyces cerevisiae, complexed with cobalt | | Descriptor: | COBALT (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3INC
 
 | |
2QIU
 
 | | Structure of Human Arg-Insulin | | Descriptor: | Insulin, ZINC ION | | Authors: | Sreekanth, R, Pattabhi, V, Rajan, S.S. | | Deposit date: | 2007-07-05 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural interpretation of reduced insulin activity as seen in the crystal structure of human Arg-insulin
Biochimie, 90, 2008
|
|
3ILG
 
 | |
2R36
 
 | | Crystal structure of ni human ARG-insulin | | Descriptor: | CHLORIDE ION, Insulin, NICKEL (II) ION, ... | | Authors: | Sreekanth, R, Pattabhi, V, Rajan, S.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal induced structural changes observed in hexameric insulin
Int.J.Biol.Macromol., 44, 2009
|
|
2R35
 
 | |
2R34
 
 | |
3IR0
 
 | |
1RKT
 
 | | Crystal structure of yfiR, a putative transcriptional regulator from Bacillus subtilis | | Descriptor: | UNKNOWN ATOM OR ION, protein yfiR | | Authors: | Anderson, W.F, Rajan, S.S, Yang, X, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-23 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of YfiR, an unusual TetR/CamR-type putative transcriptional regulator from Bacillus subtilis.
Proteins, 65, 2006
|
|
5CPG
 
 | | R-Hydratase PhaJ1 from Pseudomonas aeruginosa in the unliganded form | | Descriptor: | (R)-specific enoyl-CoA hydratase, GLYCEROL | | Authors: | Tsuge, T, Sato, S, Hiroe, A, Ishizuka, K, Kanazawa, H, Kanagarajan, S, Shiro, Y, Hisano, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Contribution of the Distal Pocket Residue to the Acyl-Chain-Length Specificity of (R)-Specific Enoyl-Coenzyme A Hydratases from Pseudomonas spp.
Appl.Environ.Microbiol., 81, 2015
|
|
6J05
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | ARSENIC, SODIUM ION, Transcriptional regulator ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
6J0E
 
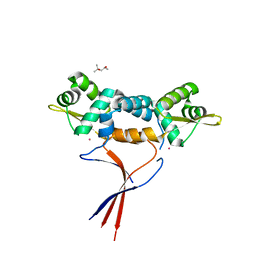 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ARSENIC, Arsenic responsive repressor ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
3F72
 
 | | Crystal Structure of the Staphylococcus aureus pI258 CadC Metal Binding Site 2 Mutant | | Descriptor: | Cadmium efflux system accessory protein, SODIUM ION | | Authors: | Kandegedara, A, Thiyagarajan, S, Kondapalli, K.C, Stemmler, T.L, Rosen, B.P. | | Deposit date: | 2008-11-07 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Role of bound Zn(II) in the CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor.
J.Biol.Chem., 284, 2009
|
|
2HTO
 
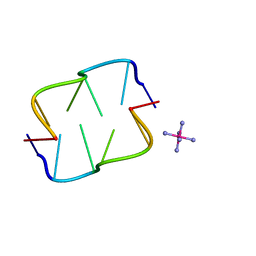 | | Ruthenium hexammine ion interactions with Z-DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), RUTHENIUM (III) HEXAAMINE ION | | Authors: | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2HTT
 
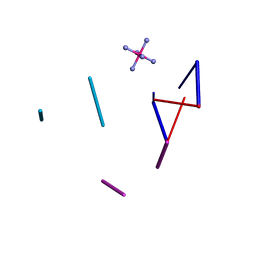 | | Ruthenium Hexammine ion interactions with Z-DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DTP*DG)-3'), ... | | Authors: | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4E3R
 
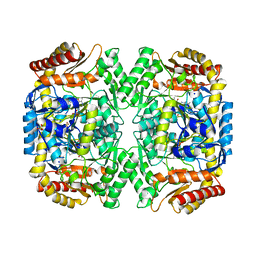 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
