1TBW
 
 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
8TUI
 
 | | Crystal structure of Fab-Lirilumab bound to KIR2DL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell immunoglobulin-like receptor 2DL3, Lirilumab Fab Heavy Chain, ... | | Authors: | Lorig-Roach, N, DuBois, R.M. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activity and specificity of the immune checkpoint inhibitor lirilumab.
Sci Rep, 14, 2024
|
|
1TC0
 
 | | Ligand Induced Conformational Shifts in the N-terminal Domain of GRP94, Open Conformation Complexed with the physiological partner ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1HB1
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1R)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOMETHYL)-2-OXOETHYL]-6-OXO-D-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
1UPW
 
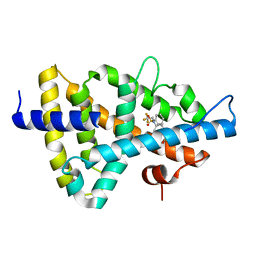 | | Crystal structure of the human Liver X receptor beta ligand binding domain in complex with a synthetic agonist | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, OXYSTEROLS RECEPTOR LXR-BETA | | Authors: | Hoerer, S, Schmid, A, Heckel, A, Budzinski, R.M, Nar, H. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Liver X Receptor Beta Ligand-Binding Domain in Complex with a Synthetic Agonist
J.Mol.Biol., 334, 2003
|
|
1TC6
 
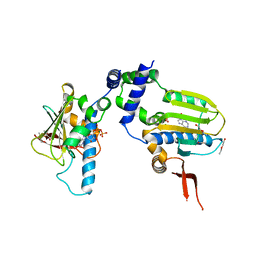 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation ADP-Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1TF6
 
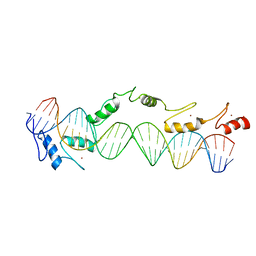 | | CO-CRYSTAL STRUCTURE OF XENOPUS TFIIIA ZINC FINGER DOMAIN BOUND TO THE 5S RIBOSOMAL RNA GENE INTERNAL CONTROL REGION | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*GP*CP*CP*TP*GP*GP*TP*TP*AP*GP*TP*AP*C P*CP*TP*GP*GP*AP* TP*GP*GP*GP*AP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*AP*GP*GP*T P*AP*CP*TP*AP*AP* CP*CP*AP*GP*GP*CP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR IIIA), ... | | Authors: | Nolte, R.T, Conlin, R.M, Harrison, S.C, Brown, R.S. | | Deposit date: | 1998-03-02 | | Release date: | 1998-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1KEZ
 
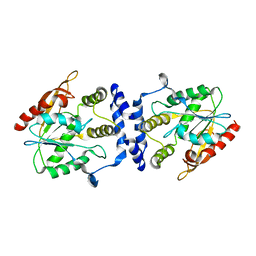 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1T61
 
 | | crystal structure of collagen IV NC1 domain from placenta basement membrane | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
1T60
 
 | | Crystal structure of Type IV collagen NC1 domain from bovine lens capsule | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
5V7C
 
 | |
1ST9
 
 | | Crystal Structure of a Soluble Domain of ResA in the Oxidised Form | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Crow, A, Acheson, R.M, Le Brun, N.E, Oubrie, A. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Redox-coupled Protein Substrate Selection by the Cytochrome c Biosynthesis Protein ResA.
J.Biol.Chem., 279, 2004
|
|
1JLO
 
 | |
1L2Y
 
 | |
1LCB
 
 | |
1UDK
 
 | | Solution Structure of Nawaprin | | Descriptor: | Nawaprin | | Authors: | Torres, A.M, Wong, H.Y, Desai, M, Moochhala, S, Kuchel, P.W, Kini, R.M. | | Deposit date: | 2003-05-01 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Identification of a Novel Family of Proteins in Snake Venoms: Purification and Structural Characterization of Nawaprin from Naja nigricollis Snake Venom
J.BIOL.CHEM., 278, 2003
|
|
1TQY
 
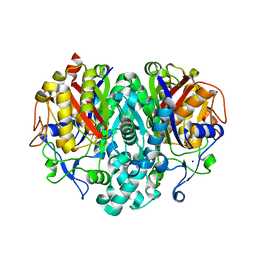 | | The Actinorhodin Ketosynthase/Chain Length Factor | | Descriptor: | ACETYL GROUP, Actinorhodin polyketide putative beta-ketoacyl synthase 1, Actinorhodin polyketide putative beta-ketoacyl synthase 2, ... | | Authors: | Keatinge-Clay, A.T, Maltby, D.A, Medzihradszky, K.F, Khosla, C, Stroud, R.M. | | Deposit date: | 2004-06-18 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An antibiotic factory caught in action.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1LCE
 
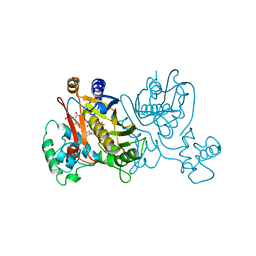 | | LACTOBACILLUS CASEI THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH DUMP AND CH2THF | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5,10-METHYLENE-6-HYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-06-22 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
1LO1
 
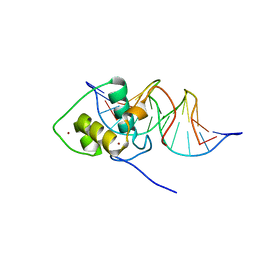 | | ESTROGEN RELATED RECEPTOR 2 DNA BINDING DOMAIN IN COMPLEX WITH DNA | | Descriptor: | 5'-D(*CP*GP*TP*GP*AP*CP*CP*TP*TP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*AP*AP*GP*GP*TP*CP*AP*CP*G)-3', Steroid hormone receptor ERR2, ... | | Authors: | Gearhart, M.D, Holmbeck, S.M.A, Evans, R.M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-05-05 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Monomeric Complex of Human Orphan Estrogen Related Receptor-2 with DNA: A Pseudo-dimer Interface Mediates Extended Half-site Recognition
J.Mol.Biol., 327, 2003
|
|
5VYO
 
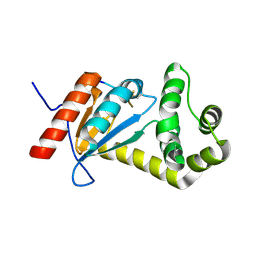 | |
1LRH
 
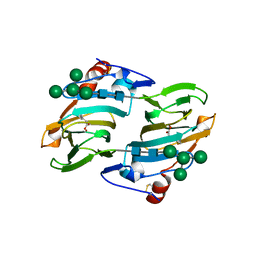 | | Crystal structure of auxin-binding protein 1 in complex with 1-naphthalene acetic acid | | Descriptor: | NAPHTHALEN-1-YL-ACETIC ACID, ZINC ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Woo, E.J, Marshall, J, Bauly, J, Chen, J.-G, Venis, M, Napier, R.M, Pickersgill, R.W. | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of auxin-binding protein 1 in complex with auxin.
EMBO J., 21, 2002
|
|
1M55
 
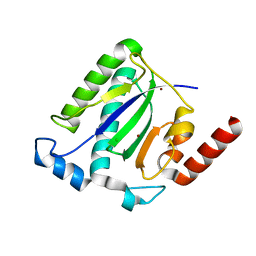 | | Catalytic domain of the Adeno Associated Virus type 5 Rep protein | | Descriptor: | CHLORIDE ION, Rep protein, ZINC ION | | Authors: | Hickman, A.B, Ronning, D.R, Kotin, R.M, Dyda, F. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep.
Mol.Cell, 10, 2002
|
|
1SE7
 
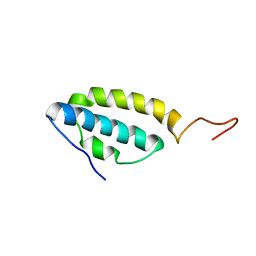 | | Solution structure of the E. coli bacteriophage P1 encoded HOT protein: a homologue of the theta subunit of E. coli DNA polymerase III | | Descriptor: | HOMOLOGUE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Chikova, A.K, Schaaper, R.M, London, R.E. | | Deposit date: | 2004-02-16 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage Like It HOT: Solution Structure of the Bacteriophage P1-Encoded HOT Protein, a Homolog of the theta Subunit of E. coli DNA Polymerase III
Structure, 12, 2004
|
|
1HKN
 
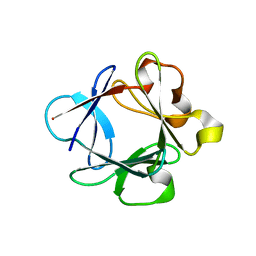 | | A complex between acidic fibroblast growth factor and 5-amino-2-naphthalenesulfonate | | Descriptor: | 5-AMINO-NAPHTALENE-2-MONOSULFONATE, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Fernandez-Tornero, C, Lozano, R.M, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Leads for Development of New Naphthalenesulfonate Derivatives with Enhanced Antiangiogenic Activity: Crystal Structure of Acidic Fibroblast Growth Factor in Complex with 5-Amino-2-Naphthalenesulfonate
J.Biol.Chem., 278, 2003
|
|
1SQG
 
 | | The crystal structure of the E. coli Fmu apoenzyme at 1.65 A resolution | | Descriptor: | SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
