3TO1
 
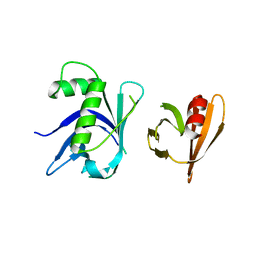 | | Two surfaces on Rtt106 mediate histone binding and chaperone activity | | Descriptor: | Histone chaperone RTT106 | | Authors: | Zunder, R.M, Antczak, A.J, Berger, J.M, Rine, J. | | Deposit date: | 2011-09-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TS3
 
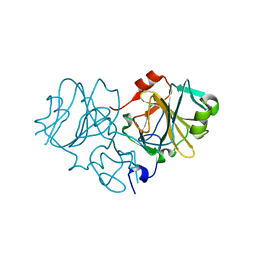 | |
2FVU
 
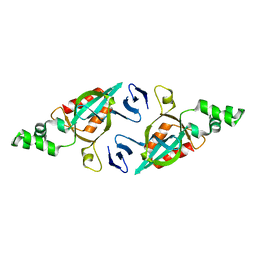 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
2FWY
 
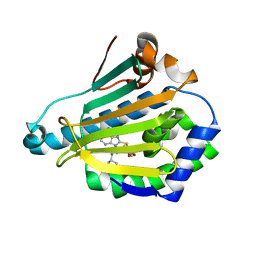 | |
2FXS
 
 | | Yeast HSP82 in complex with the novel HSP90 Inhibitor Radamide | | Descriptor: | ATP-dependent molecular chaperone HSP82, GLYCEROL, METHYL 3-CHLORO-2-{3-[(2,5-DIHYDROXY-4-METHOXYPHENYL)AMINO]-3-OXOPROPYL}-4,6-DIHYDROXYBENZOATE | | Authors: | Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
2FR1
 
 | |
2FYP
 
 | | GRP94 in complex with the novel HSP90 Inhibitor Radester amine | | Descriptor: | 2-(3-AMINO-2,5,6-TRIMETHOXYPHENYL)ETHYL 5-CHLORO-2,4-DIHYDROXYBENZOATE, Endoplasmin, PENTAETHYLENE GLYCOL, ... | | Authors: | Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibittory Ligands Adopt Different Conformations When Bound to Hsp90 or GRP94: Implications for Paralog-specific Drug Design
To be Published
|
|
3UGR
 
 | | AKR1C3 complex with indomethacin at pH 6.8 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, DIMETHYL SULFOXIDE, ... | | Authors: | Flanagan, J.U, Yosaatmadja, Y, Teague, R.M, Chai, M, Squire, C.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
3UG8
 
 | | AKR1C3 complex with indomethacin at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, INDOMETHACIN, ... | | Authors: | Flanagan, J.U, Yosaatmadja, Y, Teague, R.M, Chai, M, Squire, C.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
2FON
 
 | |
2G0B
 
 | | The structure of FeeM, an N-acyl amino acid synthase from uncultured soil microbes | | Descriptor: | FeeM, N-DODECANOYL-L-TYROSINE | | Authors: | Van Wagoner, R.M, Clardy, J. | | Deposit date: | 2006-02-11 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | FeeM, an N-Acyl Amino Acid Synthase from an Uncultured Soil Microbe: Structure, Mechanism, and Acyl Carrier Protein Binding.
Structure, 14, 2006
|
|
2G8A
 
 | | Lactobacillus casei Y261M in complex with substrate, dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, thymidylate synthase | | Authors: | Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-02 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2'-deoxyuridine 5'-monophosphate (dUMP)-binding Tyr-261
Biochemistry, 45, 2006
|
|
2GFD
 
 | | GRP94 in complex with the novel HSP90 Inhibitor Radamide | | Descriptor: | Endoplasmin, METHYL 3-CHLORO-2-{3-[(2,5-DIHYDROXY-4-METHOXYPHENYL)AMINO]-3-OXOPROPYL}-4,6-DIHYDROXYBENZOATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-03-21 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
2GF7
 
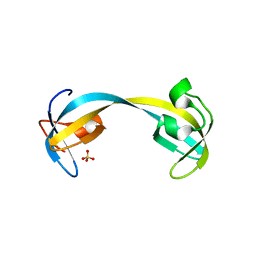 | | Double tudor domain structure | | Descriptor: | Jumonji domain-containing protein 2A, SULFATE ION | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
2FWZ
 
 | |
4ASW
 
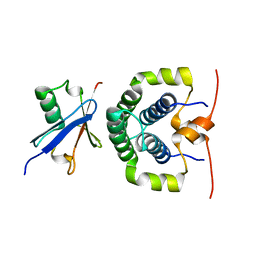 | | Structure of the complex between the N-terminal dimerisation domain of Sgt2 and the UBL domain of Get5 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2, UBIQUITIN-LIKE PROTEIN MDY2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ASV
 
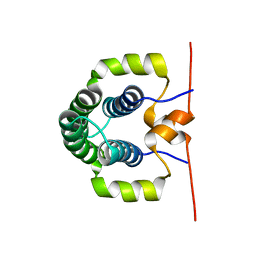 | | Solution structure of the N-terminal dimerisation domain of Sgt2 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7UK4
 
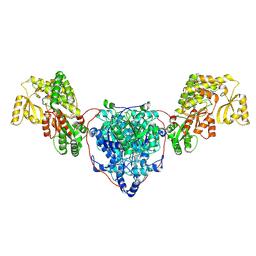 | | KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4AW0
 
 | | Human PDK1 Kinase Domain in Complex with Allosteric Compound PS182 Bound to the PIF-Pocket | | Descriptor: | 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Schulze, J.O, Busschots, K, Lopez-Garcia, L.A, Lammi, C, Stroba, A, Zeuzem, S, Piiper, A, Alzari, P.M, Neimanis, S, Arencibia, J.M, Engel, M, Biondi, R.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Substrate-Selective Inhibition of Protein Kinase Pdk1 by Small Compounds that Bind to the Pif-Pocket Allosteric Docking Site.
Chem.Biol., 19, 2012
|
|
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
7UX3
 
 | | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-05-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
6A4P
 
 | | HEWL crystals soaked in 2.5M GuHCl for 40 minutes | | Descriptor: | CHLORIDE ION, GUANIDINE, Lysozyme C, ... | | Authors: | Tushar, R, Kini, R.M, Koh, C.Y, Hosur, M.V. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray crystallographic analysis of time-dependent binding of guanidine hydrochloride to HEWL: First steps during protein unfolding.
Int. J. Biol. Macromol., 122, 2019
|
|
4GDO
 
 | | Structure of a fragment of the rod domain of plectin | | Descriptor: | Plectin | | Authors: | De Pereda, J.M, Buey, R.M, Uson, I, Sammito, M.D, De Marino, I. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
4FVP
 
 | |
