5FTM
 
 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTN
 
 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation III) | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTJ
 
 | | Cryo-EM structure of human p97 bound to UPCDC30245 inhibitor | | Descriptor: | 1-(3-(5-FLUORO-1H-INDOL-2-YL)PHENYL)PIPERIDIN-4-YL)(2-(4-ISOPROPYL-PIPERAZIN1-YL)ETHYL)-CARBAMATE, ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTK
 
 | | Cryo-EM structure of human p97 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
4V0V
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-660) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
7LGS
 
 | |
4V0U
 
 | | The crystal structure of ternary PP1G-PPP1R15B and G-actin complex | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.88 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
8G45
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G43
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G44
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(benzylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(benzylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
5ML9
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7BN9
 
 | |
4OX5
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OXD
 
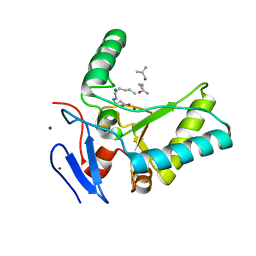 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | CHLORIDE ION, LYSINE, LdcB LD-carboxypeptidase, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
6DO1
 
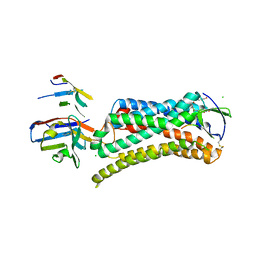 | | Structure of nanobody-stabilized angiotensin II type 1 receptor bound to S1I8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-like peptide S1I8, ... | | Authors: | Wingler, L.M, McMahon, C, Staus, D.P, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2018-06-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody.
Cell, 176, 2019
|
|
6DDA
 
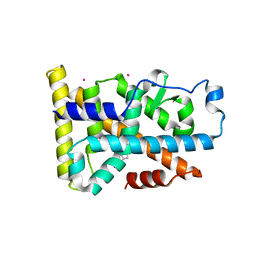 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | Descriptor: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
4OZT
 
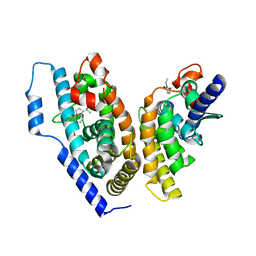 | | crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (PonA crystal) | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, N-ETHYLMALEIMIDE, ... | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-19 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6DQL
 
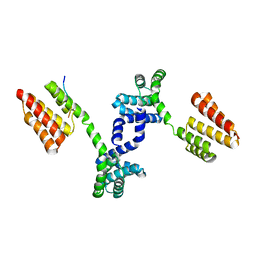 | | Crystal structure of Regulator of Proteinase B RopB complexed with SIP | | Descriptor: | Regulator of Proteinase B RopB, SpeB-inducing peptide (SIP) | | Authors: | Do, H, Makthal, N, VanderWal, A.R, Olsen, R.J, Musser, J.M, Kumaraswami, M. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway.
Nat Commun, 10, 2019
|
|
7D5G
 
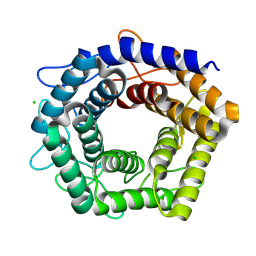 | |
5MN2
 
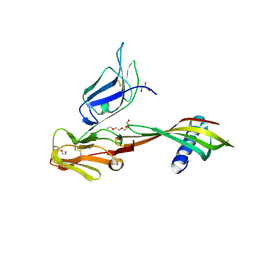 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5N11
 
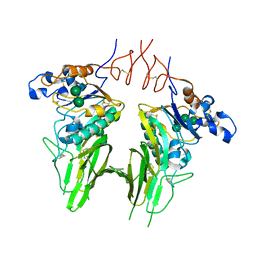 | | Crystal structure of Human beta1-coronavirus OC43 NL/A/2005 Hemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Bakkers, M.J.G, Feitsma, L.J, de Groot, R.J, Huizinga, E.G. | | Deposit date: | 2017-02-04 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity.
Cell Host Microbe, 21, 2017
|
|
7FAB
 
 | |
6ZLM
 
 | | Dihydrolipoyllysine-residue acetyltransferase component of fungal pyruvate dehydrogenase complex with protein X bound | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, Pyruvate dehydrogenase X component | | Authors: | Forsberg, B.O, Aibara, S, Howard, R.J, Mortezaei, N, Lindahl, E. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
5CQM
 
 | | GTB mutant with mercury - E303C | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
