3RKB
 
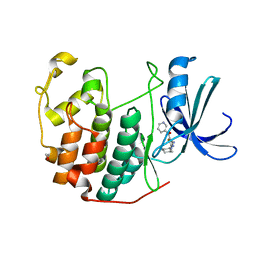 | | CDK2 in complex with inhibitor RC-2-73 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(cyclohexylamino)-1,3-thiazol-5-yl](pyridin-3-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
1A8R
 
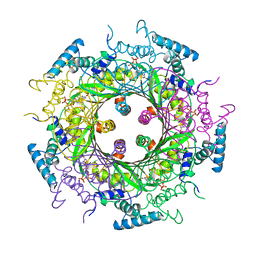 | | GTP CYCLOHYDROLASE I (H112S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-27 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of pteridines. Reaction mechanism of GTP cyclohydrolase I.
J.Mol.Biol., 326, 2003
|
|
3BYX
 
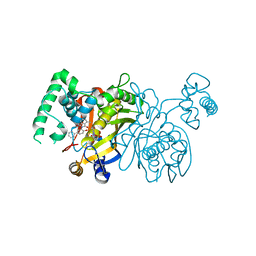 | | Lactobacillus CASEI Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative C00 in Multiple Binding Modes | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(4-hydroxybiphenyl-3-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Mangani, S, Cancian, L, Costi, M.P, Luciani, R, Ferrari, S. | | Deposit date: | 2008-01-16 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
3S0O
 
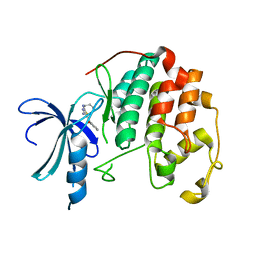 | | CDK2 in complex with inhibitor RC-1-138 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](pyridin-2-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-05-13 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3C06
 
 | | Lactobacillus CASEI Thymidylate Synthase Ternary Complex With DUMP and the Phtalimidic Derivative 14C in Multiple Binding Modes-Mode 1 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(2-chloropyridin-4-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Mangani, S, Cancian, L, Costi, M.P, Luciani, R, Ferrari, S. | | Deposit date: | 2008-01-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
7LJ9
 
 | |
1AQM
 
 | | ALPHA-AMYLASE FROM ALTEROMONAS HALOPLANCTIS COMPLEXED WITH TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 1997-07-31 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor.
Protein Sci., 7, 1998
|
|
1AQX
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH MEISENHEIMER COMPLEX | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
4CCK
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II) and N-oxalylglycine (NOG) | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
5K7F
 
 | | Crystal structure of apo AibR | | Descriptor: | ACETATE ION, Transcriptional regulator, TetR family | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
4CCN
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 L299C/C300S) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-2) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
6EKV
 
 | |
3BZ0
 
 | | Lactobacillus Casei Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative C00 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(4-hydroxybiphenyl-3-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Cancian, L, Costi, M.P, Ferrari, S, Luciani, R, Mangani, S. | | Deposit date: | 2008-01-17 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
6ER6
 
 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes7/Imdes7 | | Descriptor: | Endonuclease colEdes7, immunity Imdes7 | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
2LS5
 
 | | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
6EQZ
 
 | | A MamC-MIC insertion in MBP scaffold at position K170 | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Zarivach, R. | | Deposit date: | 2017-10-16 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | The importance of the helical structure of a MamC-derived magnetite-interacting peptide for its function in magnetite formation.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1HKF
 
 | | The three dimensional structure of NK cell receptor Nkp44, a triggering partner in natural cytotoxicity | | Descriptor: | NK CELL ACTIVATING RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-03-10 | | Release date: | 2003-06-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Three-Dimensional Structure of the Human Nk Cell Receptor Nkp44, a Triggering Partner in Natural Cytotoxicity
Structure, 11, 2003
|
|
5AP7
 
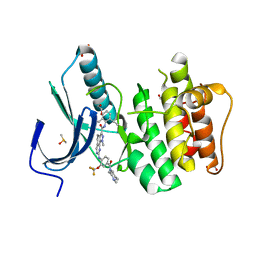 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, MONOPOLAR SPINDLE KINASE 1, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.M, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
1AQW
 
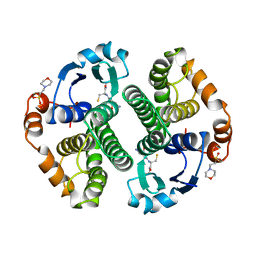 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
3C0A
 
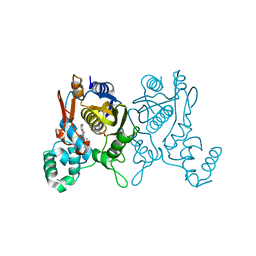 | | Lactobacillus CASEI Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative 14C in Multiple Binding Modes-Mode 2 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(2-chloropyridin-4-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Mangani, S, Cancian, L, Costi, M.P, Luciani, R, Ferrari, S. | | Deposit date: | 2008-01-19 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
1E1J
 
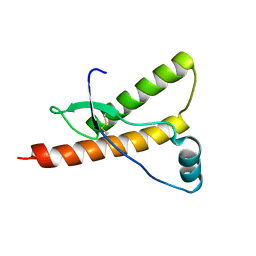 | | Human prion protein variant M166V | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-09 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DX0
 
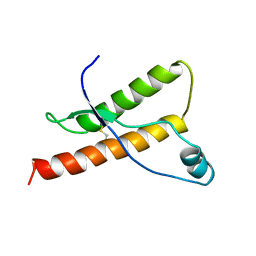 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
