7Z0A
 
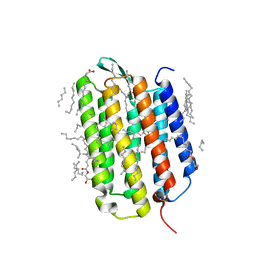 | | Crystal structure of the ground state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1MJX
 
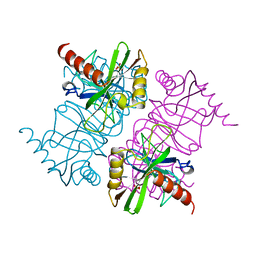 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE MUTANT D65N | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Harutyunyan, E.H, Avaeva, S.M, Huber, R. | | Deposit date: | 1997-02-08 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structures of mutant forms of E. coli inorganic pyrophosphatase with Asp-->Asn single substitution in positions 42, 65, 70, and 97.
Biochemistry Mosc., 63, 1998
|
|
7Z0D
 
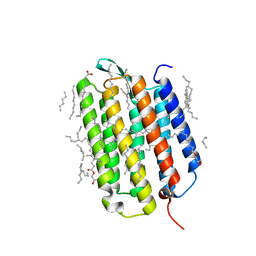 | | Crystal structure of the L state of bacteriorhodopsin at 1.20 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4FE5
 
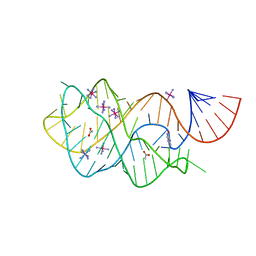 | | Crystal structure of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine.
Nature, 432, 2004
|
|
1WXH
 
 | | E.coli NAD Synthetase, NAD | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
7Z0C
 
 | | Crystal structure of the K state of bacteriorhodopsin at 1.53 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4OYF
 
 | | Crystal structure of GLTPH R397A IN Sodium-bound state | | Descriptor: | GLUTAMATE SYMPORT PROTEIN, SODIUM ION | | Authors: | Boudker, O, Oh, S, Verdon, G, Serio, R. | | Deposit date: | 2014-02-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Elife, 3, 2014
|
|
1ANU
 
 | | COHESIN-2 DOMAIN OF THE CELLULOSOME FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | COHESIN-2 | | Authors: | Shimon, L.J.W, Yaron, S, Shoham, Y, Lamed, R, Morag, E, Bayer, E.A, Frolow, F. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A cohesin domain from Clostridium thermocellum: the crystal structure provides new insights into cellulosome assembly.
Structure, 5, 1997
|
|
7Z0E
 
 | | Crystal structure of the M state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4ZKL
 
 | | Crystal structure of human histidine triad nucleotide-binding protein 1 (hHINT1) complexed with JB419 (AP4A analog) | | Descriptor: | (2R,3R,4S,5R)-2-(6-aminopurin-9-yl)-5-[2-[[(2S)-3-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-2-oxidanyl-propoxy]-sulfanyl-phosphoryl]oxyethyl]oxolane-3,4-diol, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Kaczmarek, R, Seda, A, Krakowiak, A, Baraniak, J, Nawrot, B. | | Deposit date: | 2015-04-30 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystallographic studies of the complex of human HINT1 protein with a non-hydrolyzable analog of Ap4A.
Int.J.Biol.Macromol., 87, 2016
|
|
2RLN
 
 | |
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
3VC6
 
 | | Crystal structure of enolase Tbis_1083(TARGET EFI-502310) FROM Thermobispora bispora DSM 43833 complexed with magnesium and formate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of enolase Tbis_1083 FROM Thermobispora bispora DSM 43833
To be Published
|
|
5EBI
 
 | | Crystal structure of a DNA-RNA chimera in complex with Ba2+ ions: a case of unusual multi-domain twinning | | Descriptor: | BARIUM ION, DNA/RNA (5'-D(*C)-R(P*G)-D(P*C)-R(P*G)-D(P*C)-R(P*G)-3') | | Authors: | Gilski, M, Drozdzal, P, Kierzek, R, Jaskolski, M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic resolution structure of a chimeric DNA-RNA Z-type duplex in complex with Ba(2+) ions: a case of complicated multi-domain twinning.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4K8P
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol | | Descriptor: | (2-ETHYLPHENYL)METHANOL, ADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with 2-ethylbenzyl alcohol
To be Published
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
4PTI
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Huber, R, Kukla, D, Ruehlmann, A, Epp, O, Formanek, H, Deisenhofer, J, Steigemann, W. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
6I9S
 
 | |
7VPC
 
 | | Neryl diphosphate synthase from Solanum lycopersicum | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Misawa, S, Takeshita, K, Sakai, N, Yamamoto, M, Kataoka, K, Nakayama, T, Takahashi, S, Yamashita, S. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based engineering of a short-chain cis-prenyltransferase to biosynthesize nonnatural all-cis-polyisoprenoids: molecular mechanisms for primer substrate recognition and ultimate product chain-length determination.
Febs J., 289, 2022
|
|
4OAK
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine-D-Alanine and copper (II) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3VC5
 
 | | Crystal structure of enolase Tbis_1083(TARGET EFI-502310) FROM Thermobispora bispora DSM 43833 complexed with phosphate | | Descriptor: | Mandelate racemase/muconate lactonizing protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of enolase Tbis_1083 FROM Thermobispora bispora DSM 43833
To be Published
|
|
3VDG
 
 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with formate and acetate | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enolase MSMEG_6132 FROM Mycobacterium smegmatis
To be Published
|
|
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
6IU5
 
 | | Crystal structure of cytoplasmic metal binding domain with zinc ions | | Descriptor: | CHLORIDE ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
7YWQ
 
 | |
