6BDU
 
 | |
3KNA
 
 | | M296I mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA | | Descriptor: | 3D polymerase, MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*GP*GP*CP*C)-3'), ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-12 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
6BV7
 
 | |
5EY7
 
 | |
6BGG
 
 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
5GR2
 
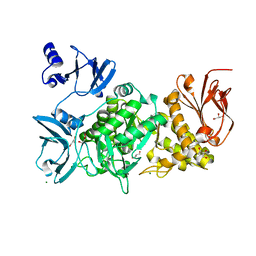 | |
6BZ9
 
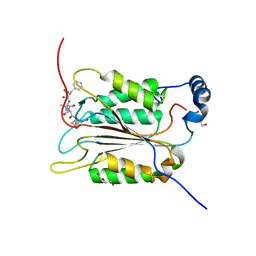 | | Crystal structure of human caspase-1 in complex with Ac-FLTD-CMK | | Descriptor: | Ac-FLTD-CMK, Caspase-1, DI(HYDROXYETHYL)ETHER | | Authors: | Xiao, T.S, Yang, J, Liu, Z, Wang, C, Yang, R. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H2Y
 
 | | human Fab 1E6 bound to fHbp variant 3 from Neisseria meningitidis serogroup B | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Veggi, D, Bianchi, F, Cozzi, R, Malito, E, Bottomley, M.J. | | Deposit date: | 2018-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cocrystal structure of meningococcal factor H binding protein variant 3 reveals a new crossprotective epitope recognized by human mAb 1E6.
Faseb J., 33, 2019
|
|
6BPI
 
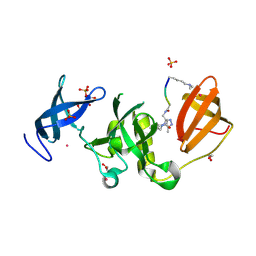 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
6H7T
 
 | | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turbe-Doan, A, Record, E, Lombard, V, Kumar, R, Henrissat, B, Levasseur, A, Garron, M.L. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The functional and structural characterization of Trichoderma reesei dehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Appl.Environ.Microbiol., 2019
|
|
6C2Y
 
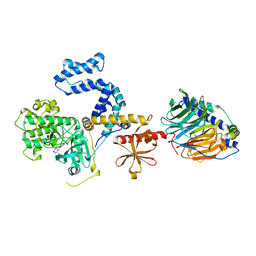 | | Human GRK2 in complex with Gbetagamma subunits and CCG257142 | | Descriptor: | (4R,5R,6S)-4-[4-fluoro-3-({[3-(methoxymethyl)-1,2,4-oxadiazol-5-yl]methyl}carbamoyl)phenyl]-N-(2H-indazol-5-yl)-6-methyl-2-oxohexahydropyrimidine-5-carboxamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Bouley, R, Tesmer, J.J.G. | | Deposit date: | 2018-01-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Utilizing a structure-based docking approach to develop potent G protein-coupled receptor kinase (GRK) 2 and 5 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6BSR
 
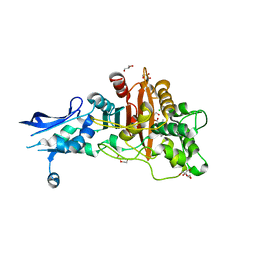 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
5GR5
 
 | |
6C1D
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C3R
 
 | |
6C42
 
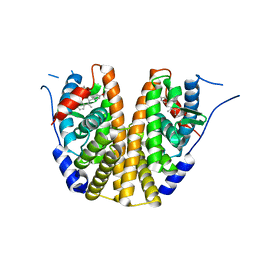 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1156 | | Descriptor: | (2R,3S,4R)-3-(4-hydroxyphenyl)-4-methyl-2-{4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}-3,4-dihydro-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallager, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2018-01-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
6BXS
 
 | |
6C9D
 
 | |
6BXR
 
 | | Crystal structure of Toxoplasma gondii Mitochondrial Association Factor 1 B (MAF1B) | | Descriptor: | BROMIDE ION, GLYCEROL, Mitochondrial association factor 1, ... | | Authors: | Parker, M.L, Ramaswamy, R, Boulanger, M.J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Toxoplasma gondii locus required for the direct manipulation of host mitochondria has maintained multiple ancestral functions.
Mol. Microbiol., 108, 2018
|
|
1BMF
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Abrahams, J.P, Leslie, A.G.W, Lutter, R, Walker, J.E. | | Deposit date: | 1996-03-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature, 370, 1994
|
|
6BXW
 
 | |
6C1I
 
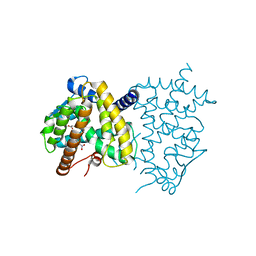 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with T0070907 | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Fuhrmann, J, Brust, R, Kojetin, D.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A structural mechanism for directing corepressor-selective inverse agonism of PPAR gamma.
Nat Commun, 9, 2018
|
|
1BOH
 
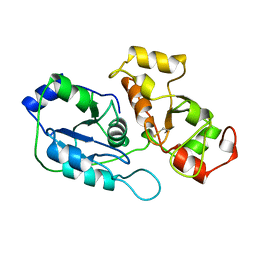 | | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
6C6T
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6CAA
 
 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
