2KVO
 
 | | Solution NMR structure of Photosystem II reaction center Psb28 protein from Synechocystis sp.(strain PCC 6803), Northeast Structural Genomics Consortium Target SgR171 | | 分子名称: | Photosystem II reaction center psb28 protein | | 著者 | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-03-22 | | 公開日 | 2010-04-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of photosystem II reaction center protein Psb28 from Synechocystis sp. Strain PCC 6803.
Proteins, 79, 2011
|
|
7MBX
 
 | | Human Cholecystokinin 1 receptor (CCK1R) Gs complex | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | 著者 | Mobbs, J.I, Belousoff, M.J, Danev, R, Thal, D.M, Sexton, P.M. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (1.95 Å) | | 主引用文献 | Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity.
Plos Biol., 19, 2021
|
|
2KW4
 
 | | Solution NMR Structure of the Holo Form of a Ribonuclease H domain from D.hafniense, Northeast Structural Genomics Consortium Target DhR1A | | 分子名称: | MAGNESIUM ION, Uncharacterized protein | | 著者 | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-03-31 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Northeast Structural Genomics Consortium Target DhR1A
To be Published
|
|
7MBY
 
 | | Human Cholecystokinin 1 receptor (CCK1R) Gq chimera (mGsqi) complex | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | 著者 | Mobbs, J.I, Belousoff, M.J, Danev, R, Thal, D.M, Sexton, P.M. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity.
Plos Biol., 19, 2021
|
|
5L71
 
 | | Crystal structure of mouse phospholipid hydroperoxide glutathione peroxidase 4 (GPx4) | | 分子名称: | 1,2-ETHANEDIOL, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | 著者 | Janowski, R, Scanu, S, Madl, T, Niessing, D. | | 登録日 | 2016-06-01 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal and solution structural studies of mouse phospholipid hydroperoxide glutathione peroxidase 4.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2KCU
 
 | | NMR solution structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107 | | 分子名称: | protein CtR107 | | 著者 | Mills, J.L, Zhang, Q, Sukumaran, D.K, Wang, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-12-29 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of an uncharacterized protein from Chlorobium tepidum. Northeast Structural Genomics target CtR107
To be Published
|
|
2KEJ
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | 分子名称: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | 著者 | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | 登録日 | 2009-01-30 | | 公開日 | 2009-05-19 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
3GK5
 
 | | Crystal structure of rhodanese-related protein (TVG0868615) from Thermoplasma volcanium, Northeast Structural Genomics Consortium Target TvR109A | | 分子名称: | Uncharacterized rhodanese-related protein TVG0868615 | | 著者 | Forouhar, F, Su, M, Seetharaman, J, Mao, L, Xiao, R, Foote, E.L, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-03-09 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of rhodanese-related protein (TVG0868615) from Thermoplasma volcanium, Northeast Structural Genomics Consortium Target TvR109A
To be Published
|
|
2KKV
 
 | | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H | | 分子名称: | Integrase | | 著者 | Mills, J.L, Wu, Y, Sukumaran, D.K, Belote, R.L, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-06-29 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H
To be Published
|
|
2KN7
 
 | | Structure of the XPF-single strand DNA complex | | 分子名称: | DNA (5'-D(*CP*AP*GP*TP*GP*GP*CP*TP*GP*A)-3'), DNA repair endonuclease XPF | | 著者 | Das, D, Folkers, G.E, van Dijk, M, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | 登録日 | 2009-08-16 | | 公開日 | 2010-08-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the XPF-ssDNA complex underscores the distinct roles of the XPF and ERCC1 helix- hairpin-helix domains in ss/ds DNA recognition
Structure, 20, 2012
|
|
2KWT
 
 | |
2KWZ
 
 | |
1NBC
 
 | |
3GYG
 
 | | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis | | 分子名称: | MAGNESIUM ION, NTD biosynthesis operon putative hydrolase ntdB | | 著者 | Nocek, B, Stein, A, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-04-03 | | 公開日 | 2009-05-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis
To be Published
|
|
1LML
 
 | | LEISHMANOLYSIN | | 分子名称: | LEISHMANOLYSIN, ZINC ION | | 著者 | Schlagenhauf, E, Etges, R, Metcalf, P. | | 登録日 | 1997-03-13 | | 公開日 | 1997-09-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The crystal structure of the Leishmania major surface proteinase leishmanolysin (gp63).
Structure, 6, 1998
|
|
1LPJ
 
 | | Human cRBP IV | | 分子名称: | Retinol-binding protein IV, cellular | | 著者 | Calderone, V, Zanotti, G, Berni, R, Folli, C. | | 登録日 | 2002-05-08 | | 公開日 | 2003-01-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ligand binding and structural analysis of a human putative cellular retinol-binding protein
J.Biol.Chem., 277, 2002
|
|
1LQJ
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | 分子名称: | URACIL-DNA GLYCOSYLASE | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4NOF
 
 | | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Polymeric immunoglobulin receptor | | 著者 | Sampathkumar, P, Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | 登録日 | 2013-11-19 | | 公開日 | 2013-12-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
1LDR
 
 | |
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | 分子名称: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | 分子名称: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | 著者 | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | 登録日 | 2003-03-28 | | 公開日 | 2004-04-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IEQ
 
 | | CRYSTAL STRUCTURE OF BARLEY BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, beta-D-glucopyranose, ... | | 著者 | Hrmova, M, DeGori, R, Fincher, G.B, Varghese, J.N. | | 登録日 | 2001-04-11 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
2KXY
 
 | | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C | | 分子名称: | Uncharacterized protein | | 著者 | Liu, Y, Lee, H, Belote, R, Ciccosanti, C, Hamilton, K, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-05-13 | | 公開日 | 2010-07-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C
To be Published
|
|
4NWC
 
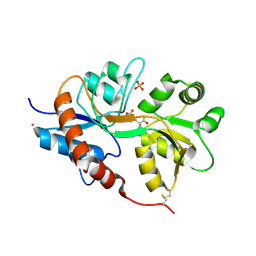 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | 分子名称: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.012 Å) | | 主引用文献 | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
2KVI
 
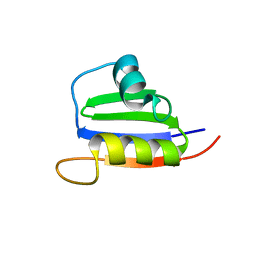 | | Structure of Nab3 RRM | | 分子名称: | Nuclear polyadenylated RNA-binding protein 3 | | 著者 | Pergoli, R, Kubicek, K, Hobor, F, Pasulka, J, Stefl, R. | | 登録日 | 2010-03-15 | | 公開日 | 2010-11-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and RNA-binding study of RNA-recognition motif of Nab3
J.Biol.Chem., 2010
|
|
