7LK0
 
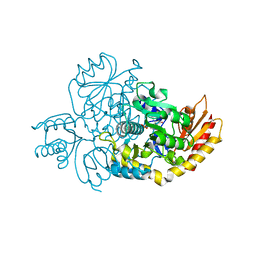 | | Ornithine Aminotransferase (OAT) cocrystallized with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) | | 分子名称: | (1R,3S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | 著者 | Butrin, A, Shen, S, Liu, D, Silverman, R. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LK1
 
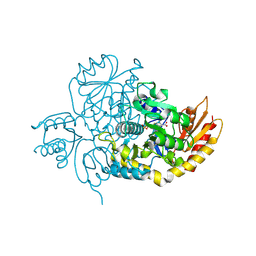 | | Ornithine Aminotransferase (OAT) with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) - 1 Hour Soaking | | 分子名称: | (1R,4R)-4-fluoro-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-2-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | 著者 | Butrin, A, Shen, S, Liu, D, Silverman, R. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
5D8Z
 
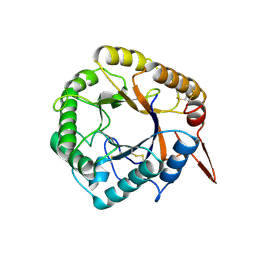 | | Structrue of a lucidum protein | | 分子名称: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, endoglucanase | | 著者 | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | 登録日 | 2015-08-18 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
5D9O
 
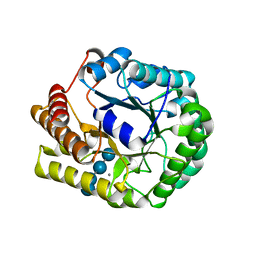 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose | | 分子名称: | B-1,4-endoglucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
7YYK
 
 | | Crystal structure of the O-fucosylated form of TSRs1-3 from the human thrombospondin 1 | | 分子名称: | 1,2-ETHANEDIOL, Thrombospondin-1, alpha-L-fucopyranose | | 著者 | Berardinelli, S.J, Eletsky, A, Valero-Gonzalez, J, Ito, A, Manjunath, R, Hurtado-Guerrero, R, Prestegard, J.R, Woods, R.J, Haltiwanger, R.S. | | 登録日 | 2022-02-18 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | O-fucosylation stabilizes the TSR3 motif in thrombospondin-1 by interacting with nearby amino acids and protecting a disulfide bond.
J.Biol.Chem., 298, 2022
|
|
7M0N
 
 | | The crystal structure of wild type PA endonuclease (A/Vietnam/1203/2004) in complex with Raltegravir | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide, ... | | 著者 | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | 登録日 | 2021-03-11 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7LXK
 
 | |
6DM3
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein | | 分子名称: | RavO | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2018-06-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein
To Be Published
|
|
6DN3
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | 分子名称: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | 登録日 | 2018-06-05 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
7S1W
 
 | | The AAVrh.10-glycan complex | | 分子名称: | Capsid protein VP1, beta-D-galactopyranose | | 著者 | Mietzsch, M, McKenna, R. | | 登録日 | 2021-09-02 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
8TOS
 
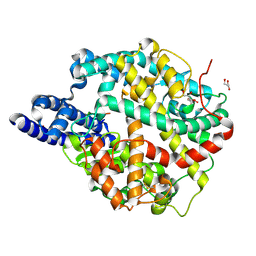 | | ACE2-peptide 6 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | 著者 | Christie, M, Payne, R. | | 登録日 | 2023-08-03 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
7OY6
 
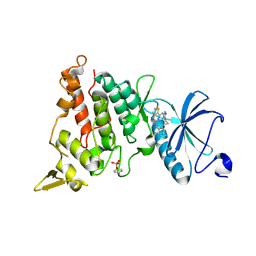 | | Crystal structure of human DYRK1A in complex with ARN25068 | | 分子名称: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | 著者 | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Ortega, J, Girotto, S, Cavalli, A. | | 登録日 | 2021-06-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
5DMH
 
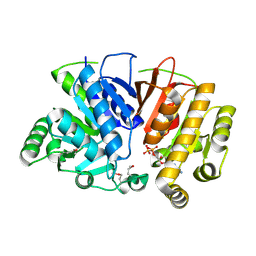 | | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein conserved in bacteria | | 著者 | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2015-09-08 | | 公開日 | 2015-10-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
To be published
|
|
7OY5
 
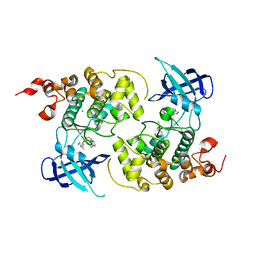 | | Crystal structure of GSK3Beta in complex with ARN25068 | | 分子名称: | CHLORIDE ION, Glycogen synthase kinase-3 beta, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | 著者 | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Giabbai, B, Storici, P, Ortega, J, Girotto, S, Cavalli, A. | | 登録日 | 2021-06-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
6XGP
 
 | | YSD1_17 major capsid protein | | 分子名称: | YSD1_17 major capsid protein | | 著者 | Grinter, R, Hardy, J.M, Dunstan, R, Lithgow, T.J, Coulibaly, F.J. | | 登録日 | 2020-06-17 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
4ZM1
 
 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - wild type | | 分子名称: | CITRIC ACID, Chain length determinant protein, MAGNESIUM ION | | 著者 | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | 登録日 | 2015-05-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
4ZRG
 
 | | Visual arrestin mutant - R175E | | 分子名称: | CARBON DIOXIDE, S-arrestin | | 著者 | Granzin, J, Stadler, A, Cousin, A, Schlesinger, R, Batra-Safferling, R. | | 登録日 | 2015-05-12 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural evidence for the role of polar core residue Arg175 in arrestin activation.
Sci Rep, 5, 2015
|
|
4G2N
 
 | | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66 | | 分子名称: | CHLORIDE ION, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding, ... | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-07-12 | | 公開日 | 2012-07-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66
To be Published
|
|
1E94
 
 | | HslV-HslU from E.coli | | 分子名称: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | 登録日 | 2000-10-07 | | 公開日 | 2000-11-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4ZZZ
 
 | | Structure of human PARP1 catalytic domain bound to an isoindolinone inhibitor | | 分子名称: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, GLYCEROL, POLY [ADP-RIBOSE] POLYMERASE 1, ... | | 著者 | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | 登録日 | 2015-04-15 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
5A3C
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | 分子名称: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | 登録日 | 2015-05-28 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
4G8S
 
 | | Crystal Structure Of a Putative Nitroreductase from Geobacter sulfurreducens PCA (Target PSI-013445) | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Nitroreductase family protein | | 著者 | Kumar, P.R, Bhosle, R, Hillerich, B, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-07-23 | | 公開日 | 2012-08-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a nitroreductase from Geobacter sulfurreducens PCA
to be published
|
|
5B1O
 
 | | DHp domain structure of EnvZ P248A mutant | | 分子名称: | Osmolarity sensor protein EnvZ | | 著者 | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
6X7H
 
 | |
4WX1
 
 | |
