2K53
 
 | | NMR solution structure of A3DK08 protein from Clostridium thermocellum: Northeast Structural Genomics Consortium Target CmR9 | | Descriptor: | A3DK08 Protein | | Authors: | Swapna, G.V.T, Huang, W, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of A3DK08 protein from Clostridium thermocellum: Northeast Structural Genomics Consortium Target CmR9
To be Published
|
|
1GV1
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1NXU
 
 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
5K8R
 
 | | Structure of human clustered protocadherin gamma B3 EC1-4 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Nicoludis, J.M, Vogt, B.E, Gaudet, R. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antiparallel protocadherin homodimers use distinct affinity- and specificity-mediating regions in cadherin repeats 1-4.
Elife, 5, 2016
|
|
5KGM
 
 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
8RPR
 
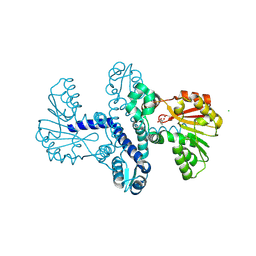 | | Crystal Structure of SgvM methyltransferase in complex with alpha-ketoleucine and Zn2+ ion | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Saleem-Batcha, R, Zou, Z, Breiltgens, J, Mueller, M, Andexer, J.N. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures and Protein Engineering of the alpha-Keto Acid C-Methyltransferases SgvM and MrsA for Rational Substrate Transfer.
Chembiochem, 2024
|
|
1L3N
 
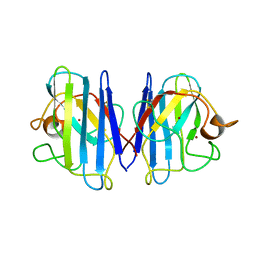 | | The Solution Structure of Reduced Dimeric Copper Zinc SOD: the Structural Effects of Dimerization | | Descriptor: | COPPER (I) ION, ZINC ION, superoxide dismutase [Cu-Zn] | | Authors: | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | Deposit date: | 2002-02-28 | | Release date: | 2002-05-08 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of reduced dimeric copper zinc superoxide dismutase. The structural effects of dimerization
Eur.J.Biochem., 269, 2002
|
|
1HB5
 
 | | quasi-atomic resolution model of bacteriophage PRD1 P3-shell, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 P3-SHELL | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
7BFY
 
 | |
2KCZ
 
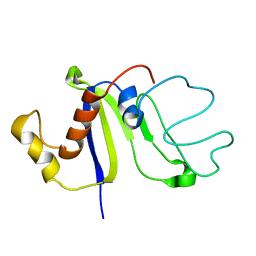 | | Solution NMR structure of the C-terminal domain of protein DR_A0006 from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR147D | | Descriptor: | Uncharacterized protein DR_A0006 | | Authors: | Mills, J.L, Ghosh, A, Garcia, E, Wang, H, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the C-Terminal Domain of Protein DR_A0006 from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR147D.
To be Published
|
|
4Z8X
 
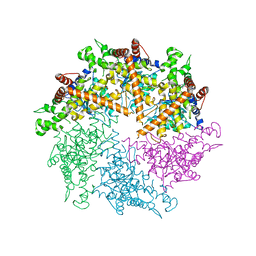 | | Truncated FtsH from A. aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, SULFATE ION, ... | | Authors: | Vostrukhina, M, Baumann, U, Schacherl, M, Bieniossek, C, Lanz, M, Baumgartner, R. | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The structure of Aquifex aeolicus FtsH in the ADP-bound state reveals a C2-symmetric hexamer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1GNZ
 
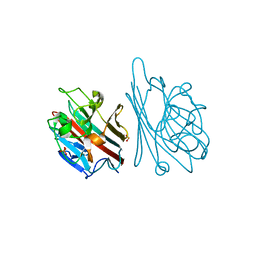 | | LECTIN I-B4 FROM GRIFFONIA SIMPLICIFOLIA (GS I-B4)METAL FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GSI-B4 ISOLECTIN, PHOSPHATE ION | | Authors: | Lescar, J, Loris, R, Mitchell, E, Gautier, C, Imberty, A. | | Deposit date: | 2001-10-11 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isolectins I-A and I-B of Griffonia (Bandeiraea) Simplicifolia. Crystal Structure of Metal-Free Gs I-B(4) and Molecular Basis for Metal Binding and Monosaccharide Specificity.
J.Biol.Chem., 277, 2002
|
|
7BQS
 
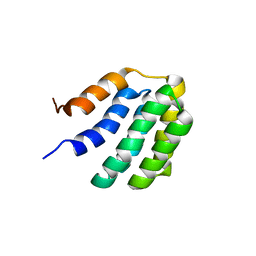 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Nomur | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Mussoc | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
1HDH
 
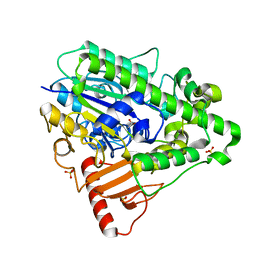 | | Arylsulfatase from Pseudomonas aeruginosa | | Descriptor: | Arylsulfatase, CALCIUM ION, SULFATE ION | | Authors: | Boltes, I, Czapinska, H, Kahnert, A, von Buelow, R, Dirks, T, Schmidt, B, von Figura, K, Kertesz, M.A, Uson, I. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 A Structure of Arylsulfatase from Pseudomonas Aeruginosa Establishes the Catalytic Mechanism of Sulfate Ester Cleavage in the Sulfatase Family.
Structure, 9, 2001
|
|
1HE4
 
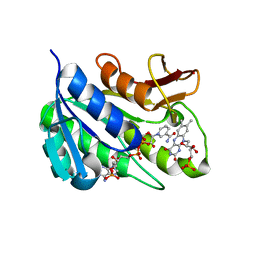 | | Human biliverdin IX beta reductase: NADP/FMN ternary complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-19 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
1OB0
 
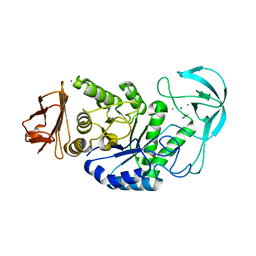 | | Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic Stabilization of Bacillus Licheniformis Alpha-Amylase Through Introduction of Hydrophobic Residues at the Surface
J.Biol.Chem., 278, 2003
|
|
4YYW
 
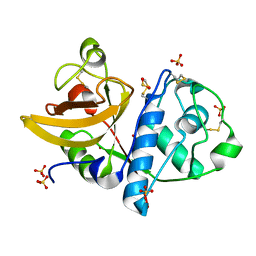 | | Ficin D2 | | Descriptor: | Ficin isoform D, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
5ICF
 
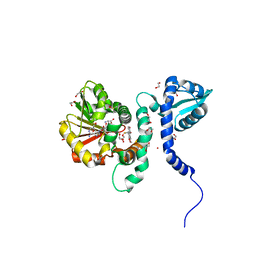 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and sanguinarine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
5ICV
 
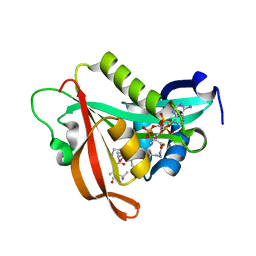 | | Crystal structure of human NatF (hNaa60) bound to a bisubstrate analogue | | Descriptor: | MET-LYS-ALA-VAL-LIG, N-alpha-acetyltransferase 60, [5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)furan-2-yl]methyl (3R)-4-{[3-({(E)-2-[(2,2-dihydroxyethyl)sulfanyl]ethenyl}amino)-3-oxopropyl]amino}-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
7BJ4
 
 | | Inulosucrase from Halalkalicoccus jeotgali bound to kestose | | Descriptor: | Levansucrase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
1HDF
 
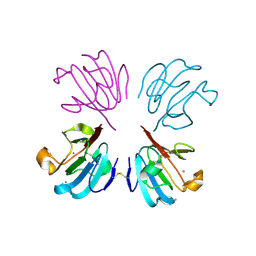 | | Evolution of the eye lens beta-gamma-crystallin domain fold | | Descriptor: | CALCIUM ION, SPHERULIN 3A | | Authors: | Clout, N.J, Kretschmar, M, Jaenicke, R, Slingsby, C. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Calcium-Loaded Spherulin 3A Dimer Sheds Light on the Evolution of the Eye Lens Betagamma-Crystallin Domain Fold
Structure, 9, 2001
|
|
2KG4
 
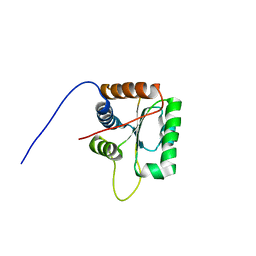 | | Three-dimensional structure of human Gadd45alpha in solution by NMR | | Descriptor: | Growth arrest and DNA-damage-inducible protein GADD45 alpha | | Authors: | Sanchez, R, Pantoja-Uceda, D, Prieto, J, Diercks, T, Campos-Olivas, R, Blanco, F.J. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase
J.Biol.Chem., 285, 2010
|
|
7BJ5
 
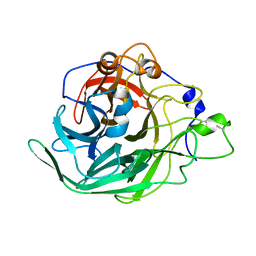 | | Inulosucrase from Halalkalicoccus jeotgali | | Descriptor: | Levansucrase | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
4WKF
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
