8F28
 
 | | Lysozyme Structures from Single-Entity Crystallization Method NanoAC | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yang, R, Sankaran, B, Ogbonna, E, Wang, G. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Single-Entity Method for Actively Controlled Nucleation and High-Quality Protein Crystal Synthesis.
Anal.Chem., 95, 2023
|
|
7A0M
 
 | | TSC1 N-terminal domain | | Descriptor: | SULFATE ION, TSC1 N-terminal domain | | Authors: | Zech, R, Kiontke, S, Kuemmel, D. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-26 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation.
Mol.Cell, 81, 2021
|
|
6ZVQ
 
 | | Complex between SMAD2 MH2 domain and peptide from Ski corepressor | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Mothers against decapentaplegic homolog 2, ... | | Authors: | Purkiss, A.G, Kjaer, S, George, R, Hill, C.S. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mutations in SKI in Shprintzen-Goldberg syndrome lead to attenuated TGF-beta responses through SKI stabilization.
Elife, 10, 2021
|
|
5HYM
 
 | | 3-Hydroxybenzoate 6-hydroxylase from Rhodococcus jostii in complex with phosphatidylinositol | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Phosphatidylinositol, ... | | Authors: | Orru, R, Montersino, S, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxybenzoate 6-Hydroxylase from Rhodococcus jostii RHA1 Contains a Phosphatidylinositol Cofactor.
Front Microbiol, 8, 2017
|
|
6XUN
 
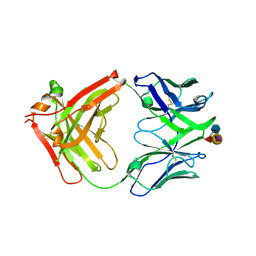 | | Ab 5b1 bound to CA19-9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
5HT7
 
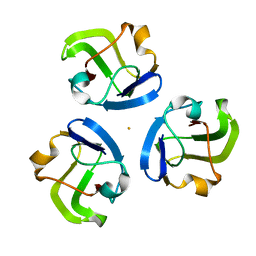 | |
6EOP
 
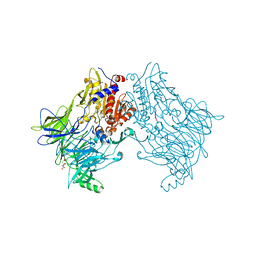 | | DPP8 - SLRFLYEG, space group 20 | | Descriptor: | CALCIUM ION, CITRATE ANION, Dipeptidyl peptidase 8, ... | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7YZ4
 
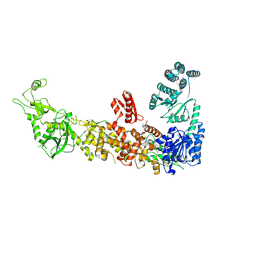 | | Mouse endoribonuclease Dicer (composite structure) | | Descriptor: | Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
6EOT
 
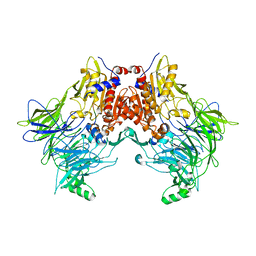 | | DPP8 - SLRFLYEG, space group 19 | | Descriptor: | Dipeptidyl peptidase 8, SER-LEU-ARG-PHE-LEU-TYR-GLU-GLY | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7YYM
 
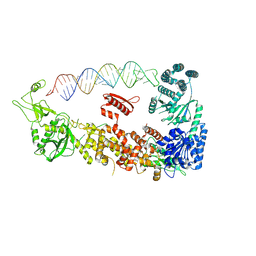 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
6RTY
 
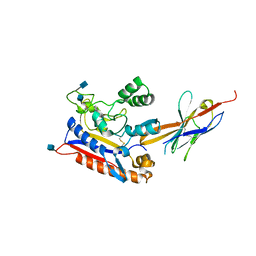 | | Crystal structure of the Patched ectodomain in complex with nanobody NB64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Llama-derived nanobody NB64, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
7YYN
 
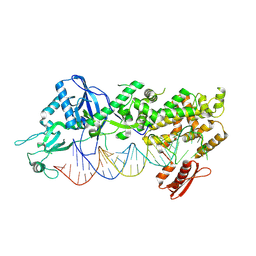 | | Mammalian Dicer in the dicing state with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Isoform 2 of Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
1OPX
 
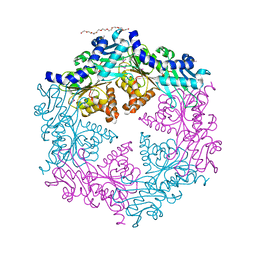 | | Crystal structure of the traffic ATPase (HP0525) of the Helicobacter pylori type IV secretion system bound by sulfate | | Descriptor: | NONAETHYLENE GLYCOL, SULFATE ION, virB11 homolog | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
6UL6
 
 | |
6UI1
 
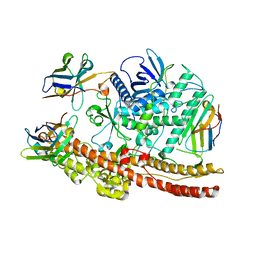 | | Crystal structure of BoNT/A-LCHn domain in complex with VHH ciA-D12, ciA-B5, and ciA-H7 | | Descriptor: | BoNT/A, ciA-B5, ciA-D12, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2019-09-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.20000863 Å) | | Cite: | Structural Insights into Rational Design of Single-Domain Antibody-Based Antitoxins against Botulinum Neurotoxins
Cell Rep, 30, 2020
|
|
6EOO
 
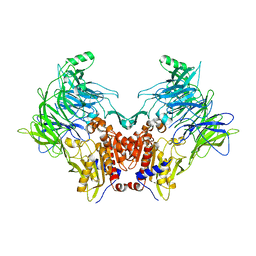 | | DPP8 - Apo, space group 20 | | Descriptor: | Dipeptidyl peptidase 8 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ADI
 
 | | KirBac3.1 W46R: role of a highly conserved tryptophan at the membrane-water interface of Kir channel | | Descriptor: | Inward rectifier potassium channel Kirbac3.1, MAGNESIUM ION, POTASSIUM ION | | Authors: | Venien-Bryan, C, Fagnen, C, De Zorzi, R, Bannwarth, L, Oubella, I, Haouz, A. | | Deposit date: | 2020-09-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Integrative Study of the Structural and Dynamical Properties of a KirBac3.1 Mutant: Functional Implication of a Highly Conserved Tryptophan in the Transmembrane Domain.
Int J Mol Sci, 23, 2021
|
|
6UC6
 
 | |
6XKS
 
 | | Crystal structure of domain A from the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) of Salmonella typhimurium | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Histidine ABC transporter substrate-binding protein HisJ | | Authors: | Romero-Romero, S, Berrocal, T, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermodynamic and kinetic analysis of the LAO binding protein and its isolated domains reveal non-additivity in stability, folding and function.
Febs J., 2023
|
|
6RTX
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6RVD
 
 | | Revised cryo-EM structure of the human 2:1 Ptch1-Shh complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | El Omari, K, Rudolf, A.F, Kowatsch, C, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6V8Z
 
 | |
4QI4
 
 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase, MtDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
5IIG
 
 | |
6EOS
 
 | | DPP8 - Apo, space group 19 | | Descriptor: | Dipeptidyl peptidase 8 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
