7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
8YHH
 
 | | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus
To Be Published
|
|
5WXC
 
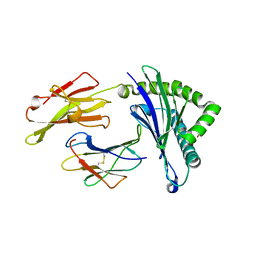 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 2) | | Descriptor: | Beta-2-microglobulin, H7-25-F, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
6LMJ
 
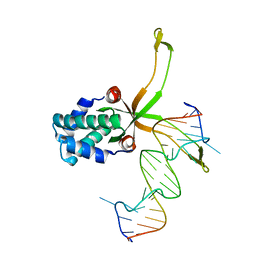 | | ASFV pA104R in complex with double-strand DNA | | Descriptor: | A104R, DNA (5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*A)-3') | | Authors: | Wang, H, Qi, J, Chai, Y, Gao, F, Liu, R. | | Deposit date: | 2019-12-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
6LMH
 
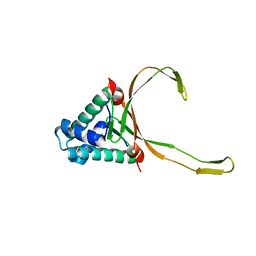 | |
8J5J
 
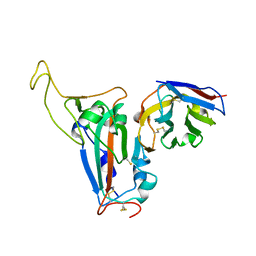 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
5WWU
 
 | | Crystal Structure of HLA-A*2402 in complex with 2009 pandemic influenza A(H1N1) virus and avian influenza A(H5N1) virus-derived peptide H1-25 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-05 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
6LXK
 
 | | Crystal structure of Z2B3 D102R Fab in complex with influenza virus neuraminidase from A/Serbia/NS-601/2014 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3-D102R Fab, Light chain of Z2B3-D102R Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.608 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXJ
 
 | | Crystal structure of human Z2B3 Fab in complex with influenza virus neuraminidase from A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Heavy chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXI
 
 | | Crystal structure of Z2B3 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3 Fab, Light chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
8YHK
 
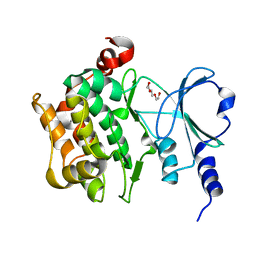 | | The Crystal Structure of P21-Activated Kinases Pak4 from Biortus | | Descriptor: | Serine/threonine-protein kinase PAK 4, TRIETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of P21-Activated Kinases Pak4 from Biortus
To Be Published
|
|
8YHI
 
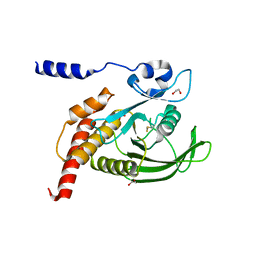 | | The Crystal Structure of SHP1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of SHP1 from Biortus.
To Be Published
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
8ZGD
 
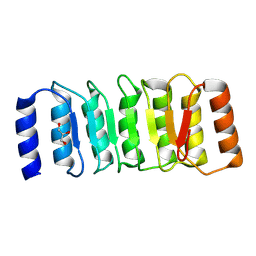 | | The Crystal Structure of the NLRP1_LRR domain from Biortus. | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Pan, W. | | Deposit date: | 2024-05-09 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the NLRP1_LRR domain from Biortus.
To Be Published
|
|
5WWJ
 
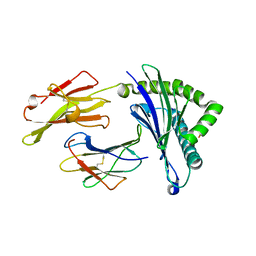 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1).
To Be Published
|
|
8ZKD
 
 | | The Crystal Structure of the RON from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Macrophage-stimulating protein receptor beta chain, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Pan, W. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the RON from Biortus.
To Be Published
|
|
6LUQ
 
 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
5WZ1
 
 | | Crystal structure of Zika virus NS5 methyltransferase bound to S-adenosyl-L-methionine | | Descriptor: | NS5 methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5WWI
 
 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1).
To Be Published
|
|
5WZ2
 
 | | Crystal structure of Zika virus NS5 methyltransferase bound to SAM and RNA analogue (m7GpppA) | | Descriptor: | NS5 MTase, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
3AL4
 
 | | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhang, W, Qi, J.X, Shi, Y, Li, Q, Yan, J.H, Gao, G.F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus
Protein Cell, 1, 2010
|
|
6KY9
 
 | | Crystal structure of ASFV dUTPase and UMP complex | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
