1XBL
 
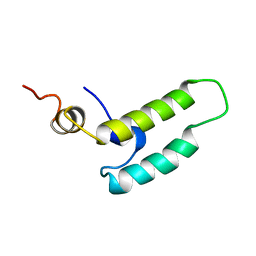 | | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | | Descriptor: | DNAJ | | Authors: | Pellecchia, M, Szyperski, T, Wall, D, Georgopoulos, C, Wuthrich, K. | | Deposit date: | 1996-10-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
J.Mol.Biol., 260, 1996
|
|
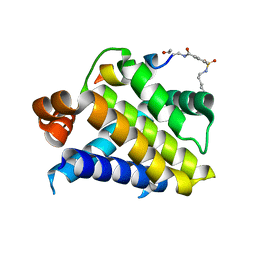
1DG4
 
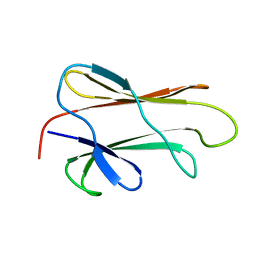 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK IN THE APO FORM | | Descriptor: | DNAK | | Authors: | Pellecchia, M, Montgomery, D.L, Stevens, S.Y, Van der Kooi, C.W, Feng, H, Gierasch, L.M, Zuiderweg, E.R.P. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate binding by the molecular chaperone DnaK.
Nat.Struct.Biol., 7, 2000
|
|
6VBX
 
 | | Crystal structure of Mcl-1 in complex with 138E12 peptide, Lys-covalent antagonist | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic peptide | | Authors: | Pellecchia, M, Perry, J.J, Kenjic, N, Assar, Z. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Structural Characterization of Lysine Covalent BH3 Peptides Targeting Mcl-1.
J.Med.Chem., 64, 2021
|
|
1BF8
 
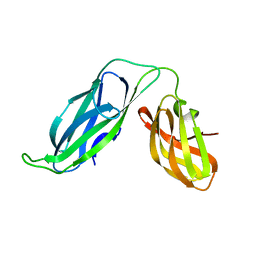 | | PERIPLASMIC CHAPERONE FIMC, NMR, 20 STRUCTURES | | Descriptor: | CHAPERONE PROTEIN FIMC | | Authors: | Pellecchia, M, Guntert, P, Glockshuber, R, Wuthrich, K. | | Deposit date: | 1998-05-28 | | Release date: | 1998-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the periplasmic chaperone FimC.
Nat.Struct.Biol., 5, 1998
|
|
8VJP
 
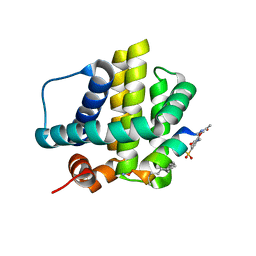 | | Histidine-covalent stapled alpha-helical peptide (155H1) targeting hMcl-1 | | Descriptor: | (4Z)-oct-4-en-1-ol, (S~1~R)-3-carbamoyl-4-methoxybenzene-1-sulfinic acid, Histidine-covalent stapled alpha-helical peptide, ... | | Authors: | Muzzarelli, K.M, Assar, Z, Alboreggia, G, Pellecchia, M. | | Deposit date: | 2024-01-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Histidine-Covalent Stapled Alpha-Helical Peptides Targeting hMcl-1.
J.Med.Chem., 67, 2024
|
|
8GH7
 
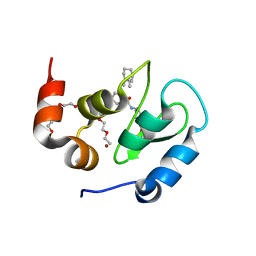 | | 142D6 bound to BIR3-XIAP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, BIR3 inhibitor MAA-CHG-PRO-ZHW, E3 ubiquitin-protein ligase XIAP, ... | | Authors: | Garza-Granados, A, McGuire, J, Baggio, C, Pellecchia, M, Pegan, S.D. | | Deposit date: | 2023-03-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a Potent and Orally Bioavailable Lys-Covalent Inhibitor of Apoptosis Protein (IAP) Antagonist.
J.Med.Chem., 66, 2023
|
|
7OFV
 
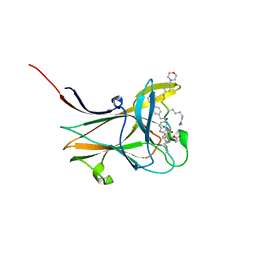 | | NMR-guided design of potent and selective EphA4 agonistic ligands | | Descriptor: | ACETATE ION, EphA4 agonist ligand, Ephrin type-A receptor 4 | | Authors: | Ganichkin, O.M, Craig, T.K, Baggio, C, Pellecchia, M. | | Deposit date: | 2021-05-05 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | NMR-Guided Design of Potent and Selective EphA4 Agonistic Ligands.
J.Med.Chem., 64, 2021
|
|
6RJP
 
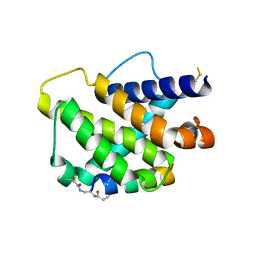 | | Bfl-1 in complex with alpha helical peptide | | Descriptor: | Bcl-2-like protein 11, Bcl-2-related protein A1 | | Authors: | Baggio, C, Gambini, L, Udompholkul, P, Salem, A.F, Hakansson, M, Jossart, J, Perry, J, Pellecchia, M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | N-locking stabilization of covalent helical peptides: Application to Bfl-1 antagonists.
Chem.Biol.Drug Des., 95, 2020
|
|
1Q5L
 
 | | NMR structure of the substrate binding domain of DnaK bound to the peptide NRLLLTG | | Descriptor: | Chaperone protein dnaK, peptide NRLLLTG | | Authors: | Stevens, S.Y, Cai, S, Pellecchia, M, Zuiderweg, E.R. | | Deposit date: | 2003-08-08 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG.
Protein Sci., 12, 2003
|
|
2A25
 
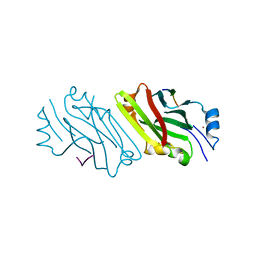 | | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP | | Descriptor: | Calcyclin-binding protein peptide, Ubiquitin ligase SIAH1, ZINC ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
2A26
 
 | | Crystal structure of the N-terminal, dimerization domain of Siah Interacting Protein | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Calcyclin-binding protein, SULFATE ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
2GJY
 
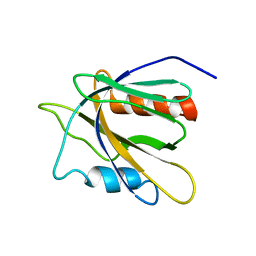 | |
2GAQ
 
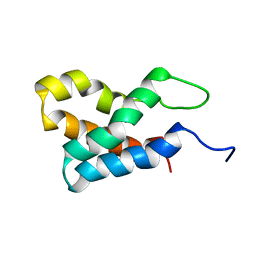 | |
4I7D
 
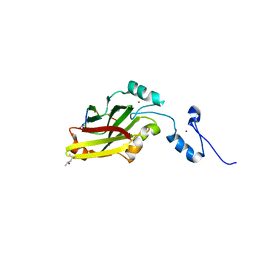 | | Siah1 bound to synthetic peptide (ACE)KLRPVAMVRP(PRK)VR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ... | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I7B
 
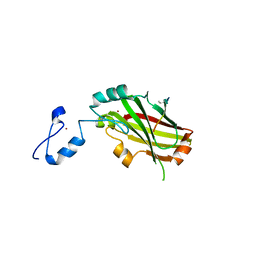 | | Siah1 bound to synthetic peptide (ACE)KLRPV(ABA)MVRPTVR | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ZINC ION | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I7C
 
 | | Siah1 mutant bound to synthetic peptide (ACE)KLRPV(23P)MVRPWVR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ... | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
2FNB
 
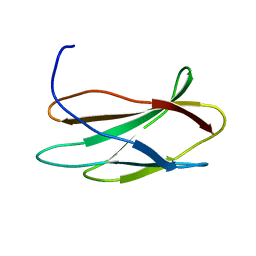 | | NMR STRUCTURE OF THE FIBRONECTIN ED-B DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Fattorusso, R, Pellecchia, M, Viti, F, Neri, P, Neri, D, Wuthrich, K. | | Deposit date: | 1998-12-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis.
Structure Fold.Des., 7, 1999
|
|
2KG5
 
 | | NMR Solution structure of ARAP3-SAM | | Descriptor: | Arf-GAP, Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Leone, M, Pellecchia, M. | | Deposit date: | 2009-03-05 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Sam domain of the lipid phosphatase Ship2 adopts a common model to interact with Arap3-Sam and EphA2-Sam.
Bmc Struct.Biol., 9, 2009
|
|
2K4P
 
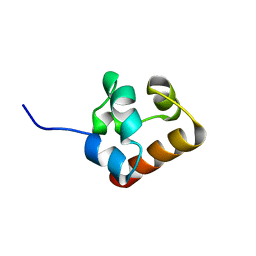 | | Solution Structure of Ship2-Sam | | Descriptor: | Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Leone, M, Pellecchia, M. | | Deposit date: | 2008-06-16 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of a Heterotypic Sam-Sam Domain Association: The Interaction between the Lipid Phosphatase Ship2 and the EphA2 Receptor.
Biochemistry, 47, 2008
|
|
2LWK
 
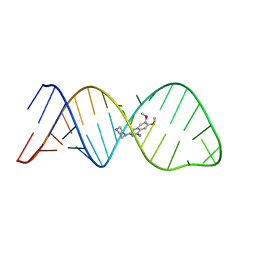 | | Solution structure of small molecule-influenza RNA complex | | Descriptor: | 6,7-dimethoxy-2-(piperazin-1-yl)quinazolin-4-amine, RNA (32-MER) | | Authors: | Lee, M.-K, Varani, G, Choi, B.-S, Pellecchia, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication.
Chem.Commun.(Camb.), 50, 2014
|
|
2LK1
 
 | |
2LK0
 
 | |
