5MG8
 
 | | Crystal structure of the S.pombe Smc5/6 hinge domain | | Descriptor: | GLYCEROL, SULFATE ION, Structural maintenance of chromosomes protein 5, ... | | Authors: | Alt, A, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Specialized interfaces of Smc5/6 control hinge stability and DNA association.
Nat Commun, 8, 2017
|
|
3ZVM
 
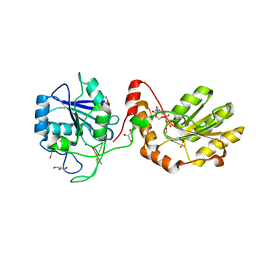 | | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*GP*TP*CP*AP*CP)-3', ACETATE ION, ... | | Authors: | Garces, F, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase.
Mol. Cell, 44, 2011
|
|
4UUH
 
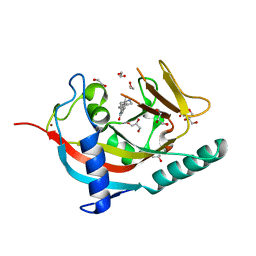 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-3-[4-(piperazin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, ... | | Authors: | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | Deposit date: | 2014-07-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1- Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
4V81
 
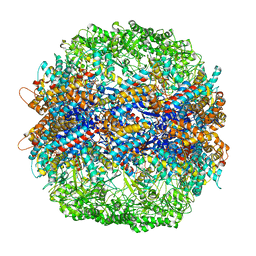 | | The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, SULFATE ION, ... | | Authors: | Dekker, C, Roe, S.M, McCormack, E.A, Beuron, F, Pearl, L.H, Willison, K.R. | | Deposit date: | 2010-10-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins.
Embo J., 30, 2011
|
|
5J3S
 
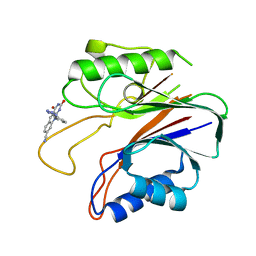 | | Crystal structure of the catalytic domain of human tyrosyl DNA phosphodiesterase 2 in complex with a small molecule inhibitor | | Descriptor: | 2,4-dioxo-10-[3-(1H-tetrazol-5-yl)phenyl]-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, Tyrosyl-DNA phosphodiesterase 2 | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
5J42
 
 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 10-(4-hydroxyphenyl)-2,4-dioxo-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, GLYCEROL, ... | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
3EU7
 
 | | Crystal Structure of a PALB2 / BRCA2 complex | | Descriptor: | 19meric peptide from Breast cancer type 2 susceptibility protein, GLYCEROL, Partner and localizer of BRCA2 | | Authors: | Oliver, A.W, Pearl, L.H. | | Deposit date: | 2008-10-09 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recruitment of BRCA2 by PALB2
Embo Rep., 10, 2009
|
|
5J3Z
 
 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-10-[3-(1H-tetrazol-5-yl)phenyl]-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, ACETATE ION, ... | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
5J3P
 
 | | Crystal structure of the catalytic domain of human tyrosyl DNA phosphodiesterase 2 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tyrosyl-DNA phosphodiesterase 2 | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
7AKO
 
 | | Crystal structure of CHK1 kinase domain in complex with a CLASPIN phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Claspin, STAUROSPORINE, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recruitment of the CHK1 DNA damage kinase by the CLASPIN scaffold protein.
Structure, 29, 2021
|
|
7AKM
 
 | | Crystal structure of CHK1 kinase domain in complex with ATPyS | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for recruitment of the CHK1 DNA damage kinase by the CLASPIN scaffold protein.
Structure, 29, 2021
|
|
1BGQ
 
 | |
7ZR5
 
 | |
7ZR0
 
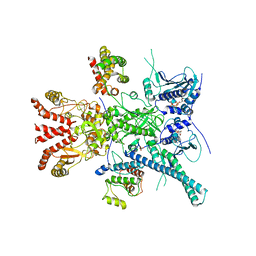 | | CryoEM structure of HSP90-CDC37-BRAF(V600E) complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Oberoi, J, Pearl, L.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | HSP90-CDC37-PP5 forms a structural platform for kinase dephosphorylation.
Nat Commun, 13, 2022
|
|
7ZR6
 
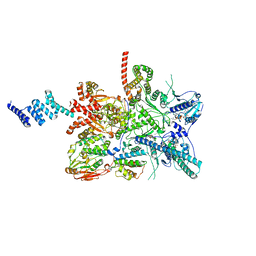 | |
5ECG
 
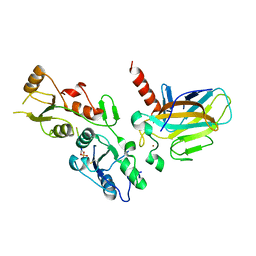 | | Crystal structure of the BRCT domains of 53BP1 in complex with p53 and H2AX-pSer139 (gammaH2AX) | | Descriptor: | Cellular tumor antigen p53, SEP-GLN-GLU-TYR, Tumor suppressor p53-binding protein 1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATM Localization and Heterochromatin Repair Depend on Direct Interaction of the 53BP1-BRCT2 Domain with gamma H2AX.
Cell Rep, 13, 2015
|
|
8AG3
 
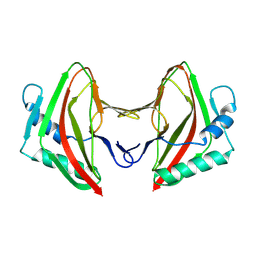 | |
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
3ZDI
 
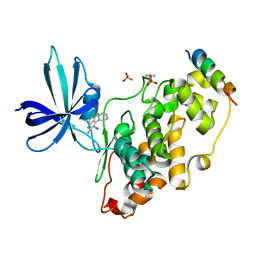 | | Glycogen Synthase Kinase 3 Beta complexed with Axin Peptide and Inhibitor 7d | | Descriptor: | 3,6-Diamino-4-(2-chlorophenyl)thieno[2,3-b]pyridine-2,5-dicarbonitrile, AXIN-1, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Oberholzer, A.E, Pearl, L.H. | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | 3,6-Diamino-4-(2-Halophenyl)-2-Benzoylthieno(2,3-B) Pyridine-5-Carbonitriles are Selective Inhibitors of Plasmodium Falciparum Glycogen Synthase Kinase-3 (Pfgsk-3)
J.Med.Chem., 56, 2013
|
|
3ZVN
 
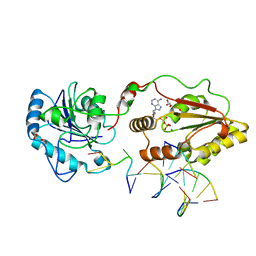 | | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase | | Descriptor: | 5'-D(*GP*TP*CP*AP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, BIFUNCTIONAL POLYNUCLEOTIDE PHOSPHATASE/KINASE, ... | | Authors: | Garces, F, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for substrate recognition by mammalian polynucleotide kinase 3' phosphatase.
Mol. Cell, 44, 2011
|
|
1GS0
 
 | |
1H8F
 
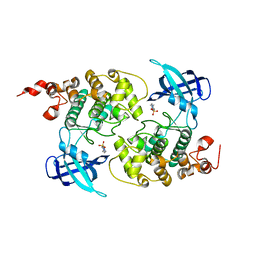 | | Glycogen Synthase Kinase 3 beta. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Dajani, R, Pearl, L.H, Roe, S.M. | | Deposit date: | 2001-02-05 | | Release date: | 2002-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Glycogen Synthase Kinase 3Beta . Structural Basis for Phosphate-Primed Substrate Specificity and Autoinhibition
Cell(Cambridge,Mass.), 105, 2001
|
|
3P23
 
 | |
