4D0A
 
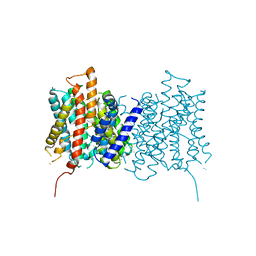 | |
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
5OYG
 
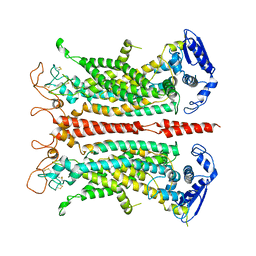 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
5OYB
 
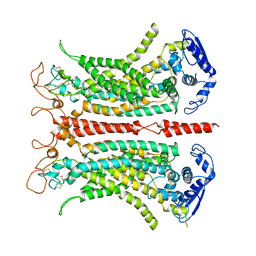 | | Structure of calcium-bound mTMEM16A chloride channel at 3.75 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
9G69
 
 | |
8QRW
 
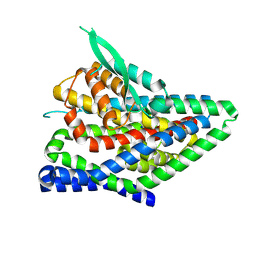 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the intermediate outward-facing state (iOFS-up) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRS
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-up) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRU
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-down) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRV
 
 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the outward-facing state (OFS) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
6RVY
 
 | |
6RVX
 
 | |
8B8G
 
 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6 | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8K
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC0
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8J
 
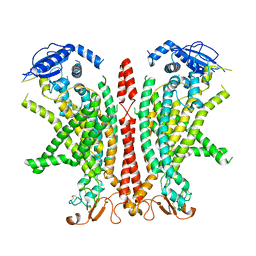 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC1
 
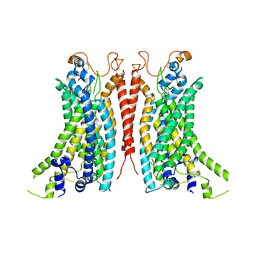 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8M
 
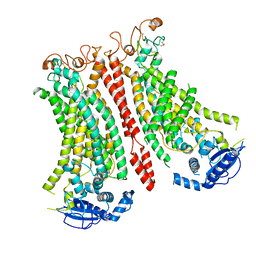 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8QRR
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.3) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRP
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.1) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRQ
 
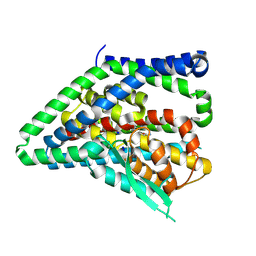 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.2) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRO
 
 | | ASCT2 trimer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8BMP
 
 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMQ
 
 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to AMP-PNP | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMS
 
 | | Cryo-EM structure of the mutant solitary ECF module 2EQ in MSP2N2 lipid nanodiscs in the ATPase closed and ATP-bound conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMR
 
 | | Cryo-EM structure of the wild-type solitary ECF module in MSP2N2 lipid nanodiscs in the ATPase open and nucleotide-free conformation | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
