5G18
 
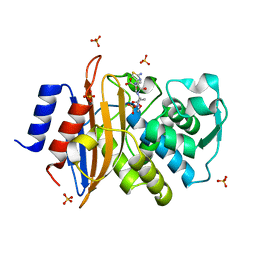 | | Direct Observation of Active-site Protonation States in a Class A beta lactamase with a monobactam substrate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BETA-LACTAMASE CTX-M-97, SULFATE ION | | Authors: | Vandavasi, V.G, Weiss, K.L, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
4N7V
 
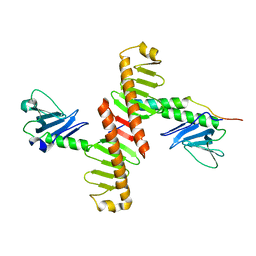 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep152 N-terminal fragment | | Descriptor: | Centrosomal protein of 152 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
6OCT
 
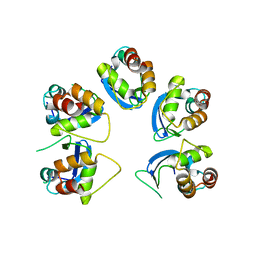 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5XJG
 
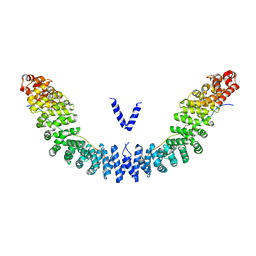 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5BW2
 
 | | X-ray crystal structure of Aplysia californica acetylcholine binding protein (Ac-AChBP) Y55W in complex with 2-Pyridin-3-yl-1-aza-bicyclo[2.2.2]octane; 2-(3-pyridyl)quinuclidine; 2-PQ (TI-4699) | | Descriptor: | (2R)-2-(pyridin-3-yl)-1-azabicyclo[2.2.2]octane, Soluble acetylcholine receptor | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-06-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
7DCK
 
 | | Crystal structure of phosphodiesterase tw9814 | | Descriptor: | Lactamase_B domain-containing protein, MANGANESE (II) ION | | Authors: | Heo, Y, Yun, J.H, Park, J.H, Park, S.B, Cha, S.S, Lee, W. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and functional identification of the uncharacterized metallo-beta-lactamase superfamily protein TW9814 as a phosphodiesterase with unique metal coordination.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7DNZ
 
 | | Cryo-EM structure of the human ABCB6 (Hemin and GSH-bound) | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kim, S, Lee, S.S, Park, J.G, Kim, J.W, Kim, S, Kim, N.J, Hong, S, Kang, J.Y, Jin, M.S. | | Deposit date: | 2020-12-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6.
Mol.Cells, 45, 2022
|
|
7DNY
 
 | | Cryo-EM structure of the human ABCB6 (coproporphyrin III-bound) | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kim, S, Lee, S.S, Park, J.G, Kim, J.W, Kim, S, Kim, N.J, Hong, S, Kang, J.Y, Jin, M.S. | | Deposit date: | 2020-12-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6.
Mol.Cells, 45, 2022
|
|
7CYU
 
 | | Crystal structure of human BAF57 HMG domain | | Descriptor: | SULFATE ION, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 | | Authors: | Heo, Y, Yun, J.H, Park, J.H, Lee, W. | | Deposit date: | 2020-09-04 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the HMG domain of human BAF57 and its interaction with four-way junction DNA.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
8GOB
 
 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
8GOA
 
 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
9JHY
 
 | | 3-Hydroxybutyryl-CoA dehydrogenase mutant (S117A) with acetoacetyl CoA | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, NAD binding domain protein, ACETOACETYL-COENZYME A | | Authors: | Yang, J.W, Jeon, H.J, Park, S.H, Jang, S.H, Park, J.A, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2024-09-10 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights and Catalytic Mechanism of 3-Hydroxybutyryl-CoA Dehydrogenase from Faecalibacterium Prausnitzii A2-165.
Int J Mol Sci, 25, 2024
|
|
9JI0
 
 | | 3-Hydroxybutyryl-CoA dehydrogenase | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, NAD binding domain protein | | Authors: | Yang, J.W, Jeon, H.J, Park, S.H, Jang, S.H, Park, J.A, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2024-09-10 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Insights and Catalytic Mechanism of 3-Hydroxybutyryl-CoA Dehydrogenase from Faecalibacterium Prausnitzii A2-165.
Int J Mol Sci, 25, 2024
|
|
9IMA
 
 | | Cryo-EM structure for the GPRC5D complexed with Talquetamab Fab | | Descriptor: | CHOLESTEROL, G-protein coupled receptor family C group 5 member D, Talquetamab Fab (anti-GPRC5D) Heavy chain, ... | | Authors: | Jeong, J, Shin, J, Park, J, Cho, Y. | | Deposit date: | 2024-07-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural Basis for the Recognition of GPRC5D by Talquetamab, a Bispecific Antibody for Multiple Myeloma.
J.Mol.Biol., 436, 2024
|
|
5CSS
 
 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
6LXX
 
 | | Frog EPDR1 with an Ir atom | | Descriptor: | CALCIUM ION, CHLORIDE ION, Ependymin-related 1, ... | | Authors: | Park, S, Park, J. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Single Soaked Iridium (IV) Ion Observed in the Frog Ependymin-Related Protein.
Bull.Korean Chem.Soc., 41, 2020
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7CLT
 
 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3OX4
 
 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 complexed with NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
6XCQ
 
 | | Erythromycin esterase EreC, mutant H289N in its closed conformation | | Descriptor: | EreC, S-1,2-PROPANEDIOL | | Authors: | Zielinski, M, Park, J, Berghuis, A.M. | | Deposit date: | 2020-06-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insights into esterase-mediated macrolide resistance.
Nat Commun, 12, 2021
|
|
3OWO
 
 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
2RVC
 
 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | Descriptor: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | Authors: | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | Deposit date: | 2015-07-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
