5JUZ
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-06-057 | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, [(R)-(2,3-dihydro-1-benzofuran-5-yl){[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methyl]phosphonic acid | | Authors: | Park, J, Magder, A, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
5JV2
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor MIT-01-055 | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, PHOSPHATE ION, ... | | Authors: | Park, J, Magder, A, Tsakos, M, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
8D37
 
 | |
8D33
 
 | |
8D3R
 
 | |
8D42
 
 | |
5KSX
 
 | | Crystal structure of human FPPS in complex with an allosteric inhibitor AM-02-072 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [[(2~{S})-2-[[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoyl]amino]phosphonic acid | | Authors: | Park, J, Matralis, A, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
6JCG
 
 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
8DTM
 
 | | Cryo-EM structure of insulin receptor (IR) bound with S597 component 2 | | Descriptor: | Insulin mimetic peptide S597 component 2, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8DTL
 
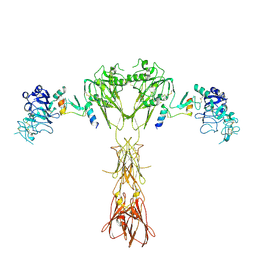 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | Descriptor: | Insulin mimetic peptide S597, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
6JCF
 
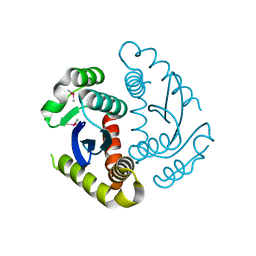 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
5JEB
 
 | |
5JA0
 
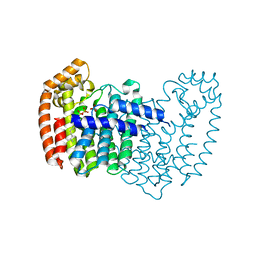 | | Crystal structure of human FPPS with allosterically bound FPP | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Zielinski, M, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human farnesyl pyrophosphate synthase is allosterically inhibited by its own product.
Nat Commun, 8, 2017
|
|
6K8C
 
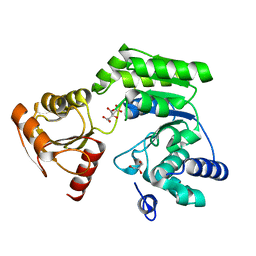 | |
6IQ6
 
 | | Crystal structure of GAPDH | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Park, J.B, Park, H.Y. | | Deposit date: | 2018-11-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Study of Monomethyl Fumarate-Bound Human GAPDH.
Mol.Cells, 42, 2019
|
|
8H53
 
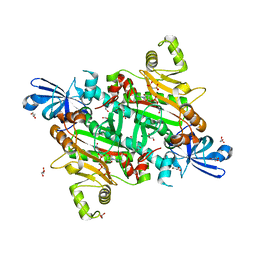 | | Human asparaginyl-tRNA synthetase in complex with asparagine-AMP | | Descriptor: | 4-AMINO-1,4-DIOXOBUTAN-2-AMINIUM ADENOSINE-5'-MONOPHOSPHATE, Asparagine--tRNA ligase, cytoplasmic, ... | | Authors: | Park, J.S, Han, B.W. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
7FCI
 
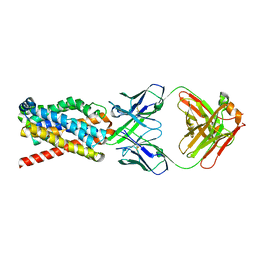 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
6L2U
 
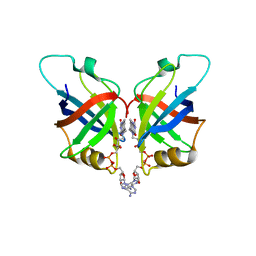 | | Soluble methane monooxygenase reductase FAD-binding domain from Methylosinus sporium. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methane monooxygenase | | Authors: | Park, J.H, Ha, S.C, Rao, Z, Yoo, H, Yoon, C, Kim, S.Y, Kim, D.S, Lee, S.J. | | Deposit date: | 2019-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5.
Dalton Trans, 50, 2021
|
|
6OAH
 
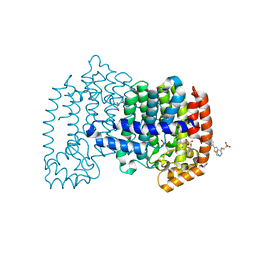 | |
6OAG
 
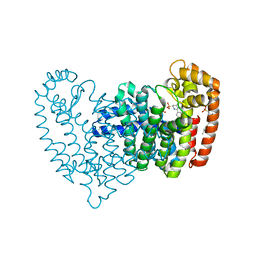 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02-82 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [(1S)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Park, J, Berghuis, A.M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
7SVW
 
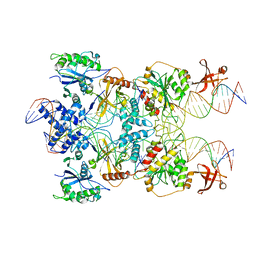 | | Strand-transfer complex of TnsB from ShCAST | | Descriptor: | MAGNESIUM ION, STC_LE_For, STC_LE_Rev1, ... | | Authors: | Park, J, Tsai, A.W.T, Kellogg, E.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Mechanistic details of CRISPR-associated transposon recruitment and integration revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SVV
 
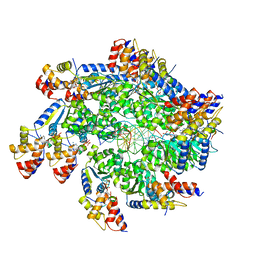 | | TnsBctd-TnsC complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Park, J, Tsai, A.W.T, Kellogg, E.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanistic details of CRISPR-associated transposon recruitment and integration revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4QY6
 
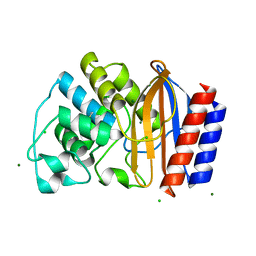 | | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations | | Descriptor: | Beta-lactamase TEM, Beta-lactamase PSE-4, CHLORIDE ION, ... | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations
To be Published
|
|
4R4R
 
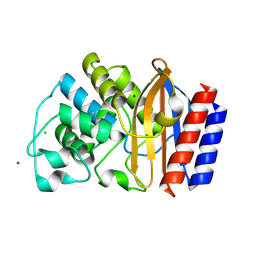 | | Crystal structure of chimeric beta-lactamase cTEM-19m at 1.2 angstrom resolution | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structural Dynamics of Engineered beta-Lactamases Vary Broadly on Three Timescales yet Sustain Native Function.
Sci Rep, 9, 2019
|
|
4R4S
 
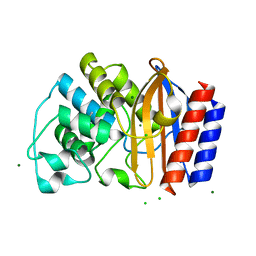 | | Crystal structure of chimeric beta-lactamase cTEM-19m at 1.1 angstrom resolution | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Structural Dynamics of Engineered beta-Lactamases Vary Broadly on Three Timescales yet Sustain Native Function.
Sci Rep, 9, 2019
|
|
