3BSF
 
 | |
7WLG
 
 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
1TX6
 
 | | trypsin:BBI complex | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Song, H.K, Park, E.Y, Kim, J.A, Kim, H.W, Kim, Y.S. | | Deposit date: | 2004-07-02 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin
J.Mol.Biol., 343, 2004
|
|
1R6L
 
 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ribonuclease PH, SULFATE ION | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
6ITC
 
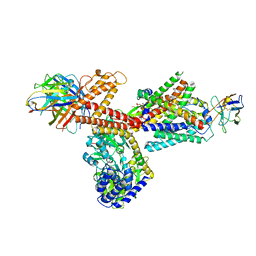 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
1R6M
 
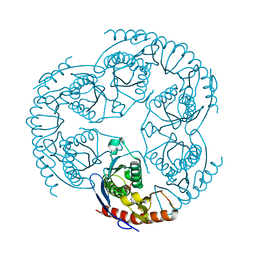 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa In Complex With Phosphate | | Descriptor: | PHOSPHATE ION, Ribonuclease PH | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
7C7C
 
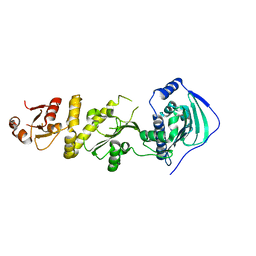 | | Crystal structure of human TRAP1 with SJT104 | | Descriptor: | 2-azanyl-9-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
7C7B
 
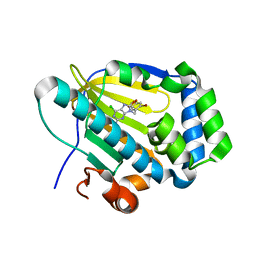 | | Crystal structure of human TRAP1 with SJT009 | | Descriptor: | 2-azanyl-9-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
5WIN
 
 | | JAK2 Pseudokinase in complex with JNJ7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5J6F
 
 | | Crystal structure of DAH7PS-CM complex from Geobacillus sp. with prephenate | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, chorismate mutase-isozyme 3, MANGANESE (II) ION, ... | | Authors: | Nazmi, A.R, Othman, M, Lang, E.J.M, Bai, Y, Allison, T.M, Panjkar, S, Arcus, V.L, Parker, E.J. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Interdomain Conformational Changes Provide Allosteric Regulation en Route to Chorismate.
J. Biol. Chem., 291, 2016
|
|
4N5V
 
 | | Alternative substrates of Mycobacterium tuberculosis anthranilate phosphoribosyl transferase | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-amino-4-fluorobenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Cookson, T.V.M, Parker, E.J, Baker, E.N, Lott, J.S. | | Deposit date: | 2013-10-10 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
3ZOK
 
 | | Structure of 3-Dehydroquinate Synthase from Actinidia chinensis in complex with NAD | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, DI(HYDROXYETHYL)ETHER, GLYCINE, ... | | Authors: | Mittelstaedt, G, Negron, L, Schofield, L.R, Marsh, K, Parker, E.J. | | Deposit date: | 2013-02-22 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterisation of dehydroquinate synthase from the New Zealand kiwifruit Actinidia chinensis.
Arch. Biochem. Biophys., 537, 2013
|
|
4UC5
 
 | | Neisseria Meningitidis DAH7PS-Phenylalanine regulated | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Heyes, L.C, Lang, E.J.M, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Calculated Pka Variations Expose Dynamic Allosteric Communication Networks.
J.Am.Chem.Soc., 138, 2016
|
|
4UMA
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3 deoxy D arabino heptulosonate 7 phosphate synthase the importance of accommodating the active site water | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4X58
 
 | | Anthranilate phosphoribosyl transferase variant N138A from Mycobacterium tuberculosis in complex with PRPP and Mg | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, D-MALATE, ... | | Authors: | Cookson, T.V.M, Evans, G.L, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
4X59
 
 | | Anthranilate phosphoribosyltransferase variant P180A from Mycobacterium tuberculosis in complex with PRPP and Mg | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Cookson, T.V.M, Evans, G.L, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
4X5A
 
 | |
4X5E
 
 | | Anthranilate phosphoribosyltransferase variant R194A from Mycobacterium tuberculosis with pyrophosphate, Mg2+ and anthranilate bound | | Descriptor: | 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Cookson, T.V.M, Evans, G.L, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
4X5C
 
 | | Anthranilate phosphoribosyltransferase variant R193L from Mycobacterium tuberculosis with pyrophosphate/PRPP and Mg2+ bound | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Cookson, T.V.M, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
4X5D
 
 | | Anthranilate phosphoribosyltransferase variant R193A from Mycobacterium tuberculosis with anthranilate bound | | Descriptor: | 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Cookson, T.V.M, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
4X5B
 
 | |
4UMC
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | L-PHOSPHOLACTATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UMB
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UCG
 
 | | NmeDAH7PS R126S variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE, ... | | Authors: | Cross, P.J, Heyes, L.C, Zhang, S, Nazmi, A.R, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Functional Unit of Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase is Dimeric.
Plos One, 11, 2016
|
|
8FEK
 
 | |
