6ZMY
 
 | | Crystal structure of hemoglobin from turkey (Meleagiris gallopova) crystallized in monoclinic form at 1.66 Angstrom resolution | | Descriptor: | Hemoglobin beta chain, Hemoglobin subunit alpha-A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pandian, R, Shobana, N, Sundaresan, S.S, Thangaraj, V, Sayed, Y, Ponnuswamy, M.N. | | Deposit date: | 2020-07-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8BHZ
 
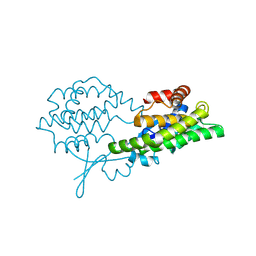 | |
1OPG
 
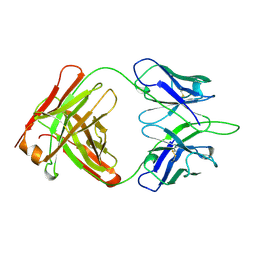 | | OPG2 FAB FRAGMENT | | Descriptor: | OPG2 FAB (HEAVY CHAIN), OPG2 FAB (LIGHT CHAIN) | | Authors: | Kodandapani, R, Veerapandian, B, Ely, K.R. | | Deposit date: | 1995-04-28 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the OPG2 Fab. An antireceptor antibody that mimics an RGD cell adhesion site.
J.Biol.Chem., 270, 1995
|
|
3BM3
 
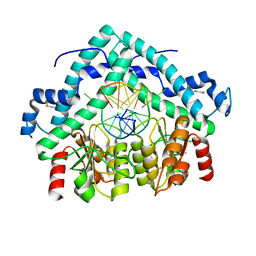 | | Restriction endonuclease PspGI-substrate DNA complex | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3'), ... | | Authors: | Szczepanowski, R.H, Carpenter, M, Czapinska, H, Tamulaitis, G, Siksnys, V, Bhagwat, A, Bochtler, M. | | Deposit date: | 2007-12-12 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A direct crystallographic demonstration that Type II restriction endonuclease PspGI flips nucleotides
To be Published
|
|
8IU6
 
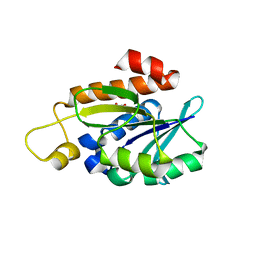 | | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Pandey, R, Tripathi, S, Lanka, A.K, Zohib, M, Pal, R.K, Arora, A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase mutant from Enterococcus faecium
To Be Published
|
|
1Z7L
 
 | |
1PUE
 
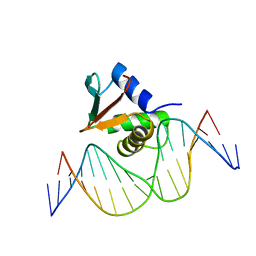 | | PU.1 ETS DOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*GP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*CP*CP*TP*TP*TP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR PU.1 (TF PU.1)) | | Authors: | Kodandapani, R, Pio, F, Ni, C.Z, Piccialli, G, Klemsz, M, McKercher, S, Maki, R.A, Ely, K.R. | | Deposit date: | 1996-07-08 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex.
Nature, 380, 1996
|
|
7Y52
 
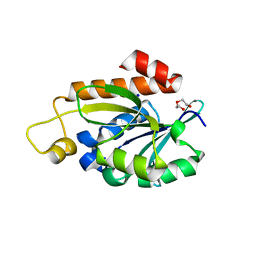 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Mundra, S, Pal, R.K, Arora, A. | | Deposit date: | 2022-06-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of structure of peptidyl-tRNA hydrolase from Enterococcus faecium and its inhibition by a pyrrolinone compound.
Int.J.Biol.Macromol., 275, 2024
|
|
4LYM
 
 | |
6CEZ
 
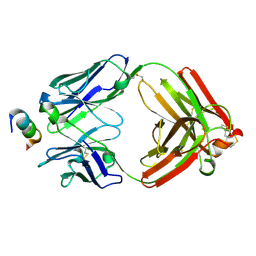 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V2 Fab 16C2 in complex with V2 peptide ConB | | Descriptor: | HIV-1 gp120 V2 Peptide Con B, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2, Light chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2 | | Authors: | Kong, X, Pan, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Select gp120 V2 domain specific antibodies derived from HIV and SIV infection and vaccination inhibit gp120 binding to alpha 4 beta 7.
PLoS Pathog., 14, 2018
|
|
6W73
 
 | |
6QM2
 
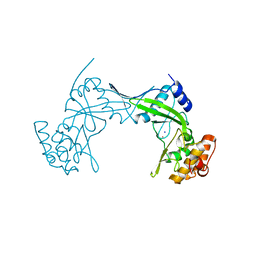 | | NlaIV restriction endonuclease | | Descriptor: | POTASSIUM ION, SODIUM ION, Type-2 restriction enzyme NlaIV | | Authors: | Czapinska, H, Siwek, W, Szczepanowski, R.H, Bujnicki, J.M, Bochtler, M, Skowronek, K. | | Deposit date: | 2019-02-01 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure and Directed Evolution of Specificity of NlaIV Restriction Endonuclease.
J.Mol.Biol., 431, 2019
|
|
8CI7
 
 | |
4XMK
 
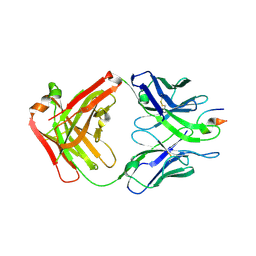 | |
4A01
 
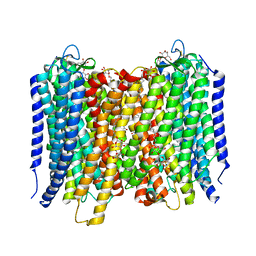 | | Crystal Structure of the H-Translocating Pyrophosphatase | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, IMIDODIPHOSPHORIC ACID, MAGNESIUM ION, ... | | Authors: | Lin, S.-M, Tsai, J.-Y, Hsiao, C.-D, Chiu, C.-L, Pan, R.-L, Sun, Y.-J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-28 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of a Membrane Embedded H1-Translocating Pyrophosphatase
Nature, 484, 2012
|
|
4XLG
 
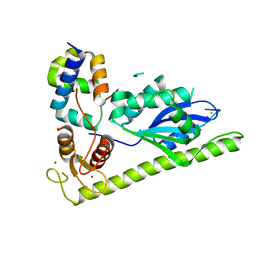 | | C. glabrata Slx1 in complex with Slx4CCD. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ... | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
6P4G
 
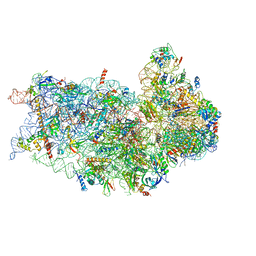 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | Descriptor: | 18S rRNA, IAPV-IRES, RACK1, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
4TX1
 
 | |
8R0S
 
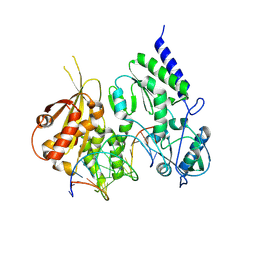 | | Structure of reverse transcriptase from Cauliflower Mosaic Virus in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*CP*GP*CP*AP*CP*TP*GP*CP*TP*GP*GP*A)-3'), Enzymatic polyprotein, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*AP*GP*UP*GP*CP*GP*UP*AP*GP*C)-3') | | Authors: | Prabaharan, C, Figiel, M, Chamera, S, Szczepanowski, R, Nowak, E, Nowotny, M. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase.
J.Biol.Chem., 300, 2024
|
|
6P4H
 
 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, IAPV-IRES, RACK1, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5I
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5N
 
 | | Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1 | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5K
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
6P5J
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
