7YD8
 
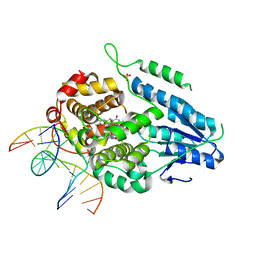 | | TR-SFX MmCPDII-DNA complex: 2 ns snapshot. Includes 2 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD6
 
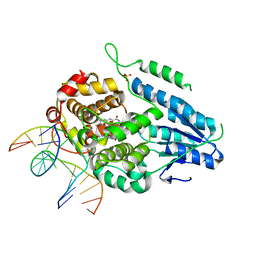 | | TR-SFX MmCPDII-DNA complex: 650 ps snapshot. Includes 650ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YDZ
 
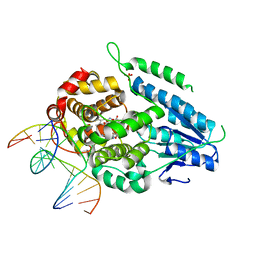 | | TR-SFX MmCPDII-DNA complex: dark state as collected in SACLA | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YE0
 
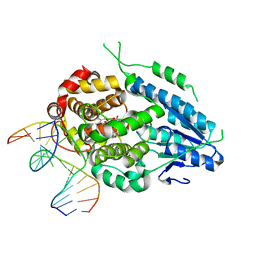 | | DF-SFX MmCPDII-DNA complex: steady state oxidized complex | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
3SNG
 
 | | X-ray structure of fully glycosylated bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-06-29 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2WIH
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-848125 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1,4,4-TETRAMETHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
3G9X
 
 | | Structure of haloalkane dehalogenase DhaA14 mutant I135F from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Lapkouski, M, Dohnalek, J, Kuta Smatanova, I. | | Deposit date: | 2009-02-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WXV
 
 | | Structure of CDK2-CYCLIN A with a Pyrazolo(4,3-h) quinazoline-3- carboxamide inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1-DIMETHYL-8-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Traquandi, G, Ciomei, M, Ballinari, D, Casale, E, Colombo, N, Croci, V, Fiorentini, F, Isacchi, A, Longo, A, Mercurio, C, Panzeri, A, Pastori, W, Pevarello, P, Volpi, D, Roussel, P, Vulpetti, A, Brasca, M.G. | | Deposit date: | 2009-11-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Potent Pyrazolo[4,3-H]Quinazoline-3-Carboxamides as Multi-Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
2WIP
 
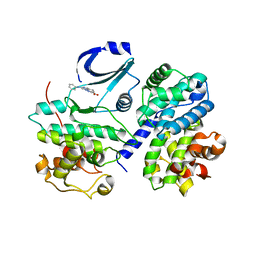 | | STRUCTURE OF CDK2-CYCLIN A COMPLEXED WITH 8-ANILINO-1-METHYL-4,5-DIHYDRO- 1H-PYRAZOLO[4,3-H] QUINAZOLINE-3-CARBOXYLIC ACID | | Descriptor: | 1-methyl-8-(phenylamino)-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxylic acid, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
5WHF
 
 | |
5WBE
 
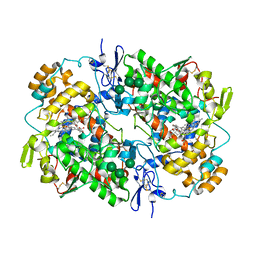 | | COX-1:MOFEZOLAC COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mofezolac, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cingolani, G, Panella, A, Perrone, M.G, Vitale, P, Smith, W.L, Scilimati, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6).
Eur J Med Chem, 138, 2017
|
|
3JB4
 
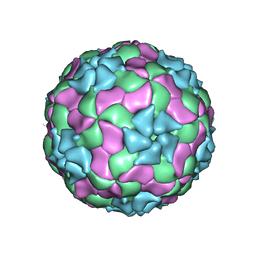 | | Structure of Ljungan virus: insight into picornavirus packaging | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Porta, C, Wenham, H, Ekstrom, J.-O, Panjwani, A, Knowles, N.J, Kotecha, A, Siebert, A, Lindberg, M, Fry, E.E, Rao, Z.H, Tuthill, T.J, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Ljungan virus provides insight into genome packaging of this picornavirus.
Nat Commun, 6, 2015
|
|
3RK4
 
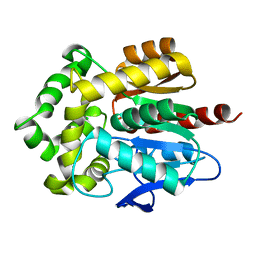 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3SK0
 
 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2011-06-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
1URW
 
 | | CDK2 IN COMPLEX WITH AN IMIDAZO[1,2-b]PYRIDAZINE | | Descriptor: | 2-[4-(N-(3-DIMETHYLAMINOPROPYL)SULPHAMOYL)ANILINO]-, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Byth, K.F, Cooper, N, Culshaw, J.D, Heaton, D.W, Oakes, S.E, Minshull, C.A, Norman, R.A, Pauptit, R.A, Tucker, J.A, Breed, J, Pannifer, A, Rowsell, S, Stanway, J.J, Valentine, A.L, Thomas, A.P. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-B]Pyridazines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2YJD
 
 | | Stapled peptide bound to Estrogen Receptor Beta | | Descriptor: | 4-(2-PROPAN-2-YLOXYBENZIMIDAZOL-1-YL)PHENOL, ESTROGEN RECEPTOR BETA, STAPLED PEPTIDE | | Authors: | Phillips, C, Roberts, L.R, Schade, M, Bent, A, Davies, N.L, Moore, R, Pannifer, A.D, Brown, D.G, Pickford, A.R, Irving, S.L. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Structure of Stapled Peptides Binding to Estrogen Receptors.
J.Am.Chem.Soc., 133, 2011
|
|
3KW8
 
 | | Two-domain laccase from Streptomyces coelicolor at 2.3 A resolution | | Descriptor: | COPPER (II) ION, FE (III) ION, Putative copper oxidase, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Duskova, J, Stepankova, A, Hasek, J, Ostergaard, L.H, Ostergaard, P.R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of laccase from Streptomyces coelicolor after soaking with potassium hexacyanoferrate and at an improved resolution of 2.3 A
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1HCZ
 
 | | LUMEN-SIDE DOMAIN OF REDUCED CYTOCHROME F AT-35 DEGREES CELSIUS | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain.
Protein Sci., 5, 1996
|
|
2XYS
 
 | | Crystal structure of Aplysia californica AChBP in complex with strychnine | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, STRYCHNINE | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2XYT
 
 | | Crystal structure of Aplysia californica AChBP in complex with d- tubocurarine | | Descriptor: | D-TUBOCURARINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
2YJA
 
 | | Stapled Peptides binding to Estrogen Receptor alpha. | | Descriptor: | ESTRADIOL, ESTROGEN RECEPTOR, STAPLED PEPTIDE | | Authors: | Phillips, C, Roberts, L.R, Schade, M, Bazin, R, Bent, A, Davies, N.L, Irving, S.L, Moore, R, Pannifer, A.D, Brown, D.G, Pickford, A.R, Scott, A, Xu, B. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design and Structure of Stapled Peptides Binding to Estrogen Receptors.
J.Am.Chem.Soc., 133, 2011
|
|
5WI0
 
 | |
5WI1
 
 | |
2YBV
 
 | | STRUCTURE OF RUBISCO FROM THERMOSYNECHOCOCCUS ELONGATUS | | Descriptor: | CHLORIDE ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL SUBUNIT | | Authors: | Terlecka, B, Wilhelmi, V, Bialek, W, Gubernator, B, Szczepaniak, A, Hofmann, E. | | Deposit date: | 2011-03-10 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase from Thermosynechococcus Elongatus
To be Published
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
