1HYS
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH A POLYPURINE TRACT RNA:DNA | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*AP*AP*AP*AP*AP*GP*TP*GP*GP*CP*TP*G)-3', 5'-R(*UP*CP*AP*GP*CP*CP*AP*CP*UP*UP*UP*UP*UP*AP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', FAB-28 MONOCLONAL ANTIBODY FRAGMENT HEAVY CHAIN, ... | | Authors: | Sarafianos, S.G, Das, K, Tantillo, C, Clark Jr, A.D, Ding, J, Whitcomb, J, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2001-01-22 | | Release date: | 2001-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA.
EMBO J., 20, 2001
|
|
1I0A
 
 | |
1HY1
 
 | |
1I6F
 
 | |
1IPS
 
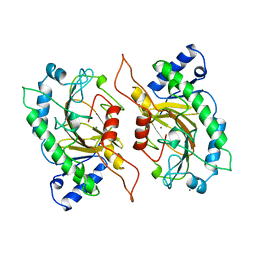 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (MANGANESE COMPLEX) | | Descriptor: | ISOPENICILLIN N SYNTHASE, MANGANESE (II) ION | | Authors: | Roach, P.L, Clifton, I.J, Fulop, V, Harlos, K, Barton, G.J, Hajdu, J, Andersson, I, Schofield, C.J, Baldwin, J.E. | | Deposit date: | 1997-03-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes.
Nature, 375, 1995
|
|
1JAB
 
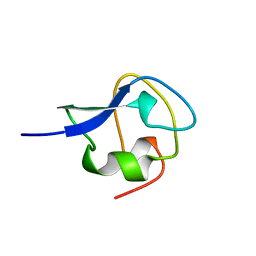 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1JP4
 
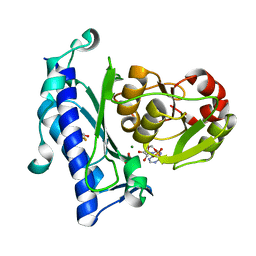 | | Crystal Structure of an Enzyme Displaying both Inositol-Polyphosphate 1-Phosphatase and 3'-Phosphoadenosine-5'-Phosphate Phosphatase Activities | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Patel, S, Yenush, L, Rodriguez, P.L, Serrano, R, Blundell, T.L. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of an enzyme displaying both inositol-polyphosphate-1-phosphatase and 3'-phosphoadenosine-5'-phosphate phosphatase activities: a novel target of lithium therapy.
J.Mol.Biol., 315, 2002
|
|
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | Descriptor: | ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2001-09-13 | | Release date: | 2002-10-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
1K62
 
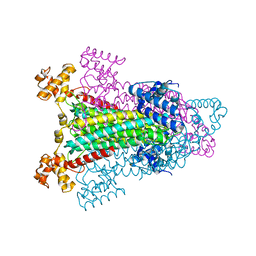 | |
1I6G
 
 | |
1HST
 
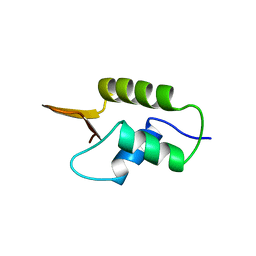 | | CRYSTAL STRUCTURE OF GLOBULAR DOMAIN OF HISTONE H5 AND ITS IMPLICATIONS FOR NUCLEOSOME BINDING | | Descriptor: | HISTONE H5 | | Authors: | Ramakrishnan, V, Finch, J.T, Graziano, V, Lee, P.L, Sweet, R.M. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of globular domain of histone H5 and its implications for nucleosome binding.
Nature, 362, 1993
|
|
1KRF
 
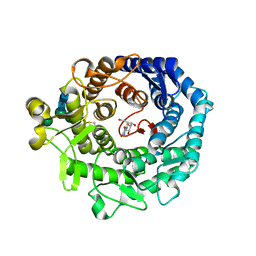 | | STRUCTURE OF P. CITRINUM ALPHA 1,2-MANNOSIDASE REVEALS THE BASIS FOR DIFFERENCES IN SPECIFICITY OF THE ER AND GOLGI CLASS I ENZYMES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KIFUNENSINE, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
1JO5
 
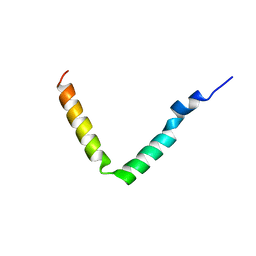 | | Rhodobacter sphaeroides Light Harvesting 1 beta Subunit in Detergent Micelles | | Descriptor: | LIGHT-HARVESTING PROTEIN B-875 | | Authors: | Sorgen, P.L, Cahill, S.M, Krueger-Koplin, R.D, Krueger-Koplin, S.T, Schenck, C.G, Girvin, M.E. | | Deposit date: | 2001-07-26 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Rhodobacter sphaeroides light-harvesting 1 beta subunit in detergent micelles.
Biochemistry, 41, 2002
|
|
1KPY
 
 | |
1KPZ
 
 | |
1KXR
 
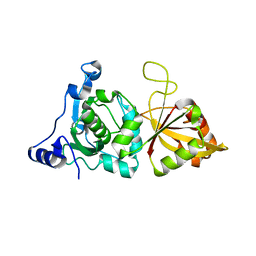 | | Crystal Structure of Calcium-Bound Protease Core of Calpain I | | Descriptor: | CALCIUM ION, thiol protease DOMAINS I AND II | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Elce, J.S, Jia, Z, Davies, P.L. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A Ca(2+) switch aligns the active site of calpain.
Cell(Cambridge,Mass.), 108, 2002
|
|
1KDE
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, 22 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
1K92
 
 | |
1KDF
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
1L0M
 
 | |
1KKT
 
 | | Structure of P. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the ER and Golgi Class I enzymes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannosyl-oligosaccharide alpha-1,2-mannosidase, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
1KP3
 
 | |
1KP2
 
 | |
1LI4
 
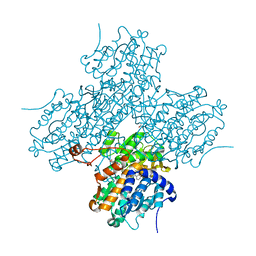 | | Human S-adenosylhomocysteine hydrolase complexed with neplanocin | | Descriptor: | 3-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-CYCLOPENTANE-1,2-DIOL, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, X, Hu, Y, Yin, D.H, Turner, M.A, Wang, M, Borchardt, R.T, Howell, P.L, Kuczera, K, Schowen, R.L. | | Deposit date: | 2002-04-17 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic strategy of S-adenosyl-L-homocysteine hydrolase: Transition-state
stabilization and the avoidance of abortive reactions
Biochemistry, 42, 2003
|
|
1LQ7
 
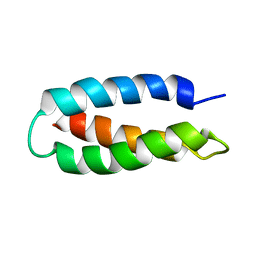 | | De Novo Designed Protein Model of Radical Enzymes | | Descriptor: | Alpha3W | | Authors: | Dai, Q.-H, Tommos, C, Fuentes, E.J, Blomberg, M, Dutton, P.L, Wand, A.J. | | Deposit date: | 2002-05-09 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a De Novo Designed Protein Model of Radical Enzymes
J.Am.Chem.Soc., 124, 2002
|
|
