6OZ1
 
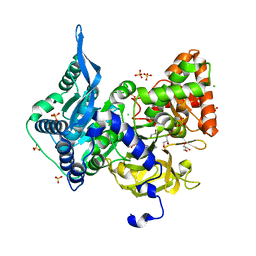 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
6FGM
 
 | | The NMR solution structure of the peptide AC12 from Hypsiboas raniceps | | Descriptor: | ALA-CYS-PHE-LEU-THR-ARG-LEU-GLY-THR-TYR-VAL-CYS | | Authors: | Popov, C.S.F.C, Simas, B.S, Goodfellow, B.J, Bocca, A.L, Andrade, P.B, Pereira, D, Valentao, P, Pereira, P.J.B, Rodrigues, J.E, Veloso Jr, P.H.H, Rezende, T.M.B. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Host-defense peptides AC12, DK16 and RC11 with immunomodulatory activity isolated from Hypsiboas raniceps skin secretion.
Peptides, 113, 2019
|
|
3KIA
 
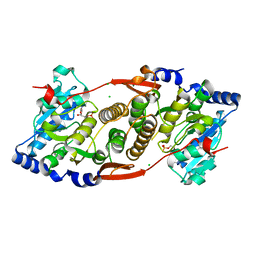 | | Crystal structure of mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | Deposit date: | 2009-11-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
1X7S
 
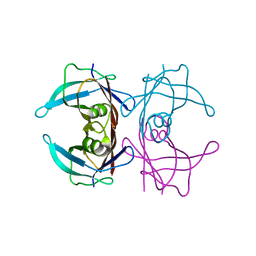 | | The X-ray crystallographic structure of the amyloidogenic variant TTR Tyr78Phe | | Descriptor: | Transthyretin | | Authors: | Neto-Silva, R.M, Macedo-Ribeiro, S, Pereira, P.J.B, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2004-08-16 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic studies of two transthyretin variants: further insights into amyloidogenesis.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1X7T
 
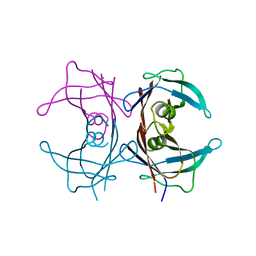 | | Structure of TTR R104H: a non-amyloidogenic variant with protective clinical effects | | Descriptor: | Transthyretin | | Authors: | Neto-Silva, R.M, Macedo-Ribeiro, S, Pereira, P.J.B, Coll, M, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2004-08-16 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of two transthyretin variants: further insights into amyloidogenesis.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6G3N
 
 | |
2BNQ
 
 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, SYNTHETIC PEPTIDE, ... | | Authors: | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, van der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines
J.Exp.Med., 201, 2005
|
|
6G7D
 
 | |
8DZV
 
 | |
1A0L
 
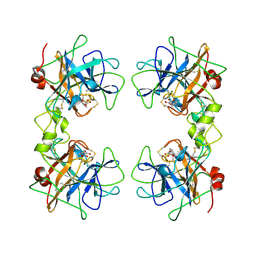 | | HUMAN BETA-TRYPTASE: A RING-LIKE TETRAMER WITH ACTIVE SITES FACING A CENTRAL PORE | | Descriptor: | (2S)-3-(4-carbamimidoylphenyl)-2-hydroxypropanoic acid, BETA-TRYPTASE | | Authors: | Pereira, P.J.B, Bergner, A, Macedo-Ribeiro, S, Huber, R, Matschiner, G, Fritz, H, Sommerhoff, C.P, Bode, W. | | Deposit date: | 1997-12-03 | | Release date: | 1999-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human beta-tryptase is a ring-like tetramer with active sites facing a central pore.
Nature, 392, 1998
|
|
2OG2
 
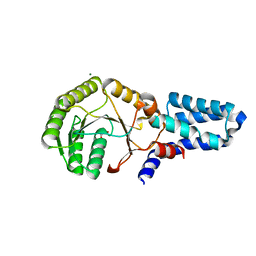 | | Crystal structure of chloroplast FtsY from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, MALONATE ION, Putative signal recognition particle receptor | | Authors: | Chartron, J, Chandrasekar, S, Ampornpan, P.J, Shan, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chloroplast Signal Recognition Particle (SRP) Receptor: Domain Arrangement Modulates SRP-Receptor Interaction.
J.Mol.Biol., 375, 2007
|
|
1CLV
 
 | | YELLOW MEAL WORM ALPHA-AMYLASE IN COMPLEX WITH THE AMARANTH ALPHA-AMYLASE INHIBITOR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE INHIBITOR), ... | | Authors: | Pereira, P.J.B, Lozanov, V, Patthy, A, Huber, R, Bode, W, Pongor, S, Strobl, S. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific inhibition of insect alpha-amylases: yellow meal worm alpha-amylase in complex with the amaranth alpha-amylase inhibitor at 2.0 A resolution.
Structure Fold.Des., 7, 1999
|
|
3JCL
 
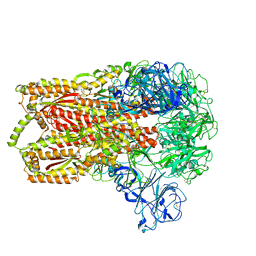 | | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Bosch, B.J, Frenz, B, Rottier, P.J.M, DiMaio, F, Rey, F.A, Veesler, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-02-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer.
Nature, 531, 2016
|
|
4WNM
 
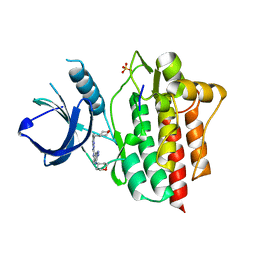 | | SYK catalytic domain in complex with a potent triazolopyridine inhibitor | | Descriptor: | N~3~-(tetrahydro-2H-pyran-4-yl)-N~6~-[5-(tetrahydro-2H-pyran-4-ylmethyl)[1,2,4]triazolo[1,5-a]pyridin-2-yl]-1H-indazole-3,6-diamine, SULFATE ION, Tyrosine-protein kinase SYK | | Authors: | Jackson, P.J. | | Deposit date: | 2014-10-13 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Triazolopyridine-Based Spleen Tyrosine Kinase Inhibitor That Arrests Joint Inflammation.
Plos One, 11, 2016
|
|
8EZR
 
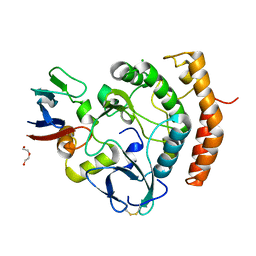 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
5M3O
 
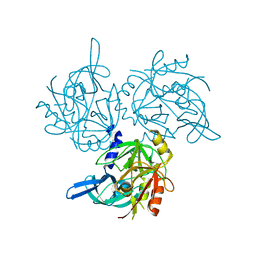 | | HTRA2 A141S mutant structure | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Serine protease HTRA2, mitochondrial | | Authors: | Merski, M, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
6G80
 
 | |
8EFZ
 
 | | Crystal structure of CcNikZ-II, apoprotein | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | Authors: | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | Deposit date: | 2022-09-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of CcNikZ-II, apoprotein
To Be Published
|
|
4Z0P
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Porebski, P.J, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
3L5I
 
 | | Crystal structure of FnIII domains of human GP130 (Domains 4-6) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-6 receptor subunit beta | | Authors: | Kershaw, N.J, Zhang, J.-G, Garrett, T.P.J, Czabotar, P.E. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
2P0C
 
 | | Catalytic Domain of the Proto-oncogene Tyrosine-protein Kinase MER | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
6H40
 
 | | High resolution structure of MeT1 from Mycobacterium hassiacum in complex with 3-methoxy-1,2-propanediol. | | Descriptor: | (2~{S})-3-methoxypropane-1,2-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pereira, P.J.B, Ripoll-Rozada, J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.053 Å) | | Cite: | Biosynthesis of mycobacterial methylmannose polysaccharides requires a unique 1-O-methyltransferase specific for 3-O-methylated mannosides.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3L5J
 
 | | Crystal structure of FnIII domains of human GP130 (Domains 4-6) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Interleukin-6 receptor subunit beta | | Authors: | Kershaw, N.J, Zhang, J.-G, Garrett, T.P.J, Czabotar, P.E. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.042 Å) | | Cite: | Crystal structure of the entire ectodomain of gp130: insights into the molecular assembly of the tall cytokine receptor complexes.
J.Biol.Chem., 285, 2010
|
|
3LL4
 
 | | Structure of the H13A mutant of Ykr043C in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Stogios, P.J, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
2A91
 
 | | Crystal structure of ErbB2 domains 1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Garrett, T.P.J, McKern, N.M, Lou, M, Elleman, T.C, Adams, T.E, Lovrecz, G.O, Kofler, M, Jorissen, R.N, Nice, E.C, Burgess, A.W. | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of a Truncated ErbB2 Ectodomain Reveals an Active Conformation, Poised to Interact with Other ErbB Receptors
Mol.Cell, 11, 2003
|
|
