6RGQ
 
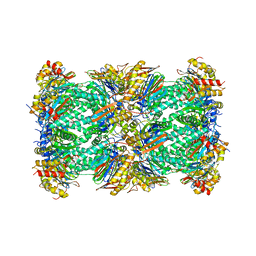 | | Human 20S Proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Toste Rego, A, da Fonseca, P.C.A. | | Deposit date: | 2019-04-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes.
Mol.Cell, 76, 2019
|
|
8PM2
 
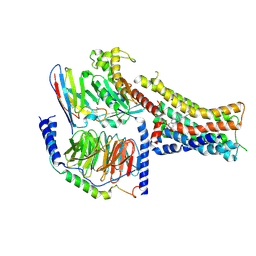 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
7QPJ
 
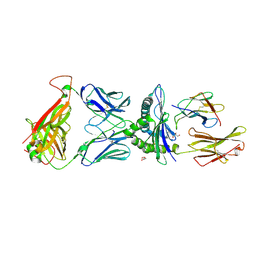 | | Crystal structure of engineered TCR (756) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
7PDX
 
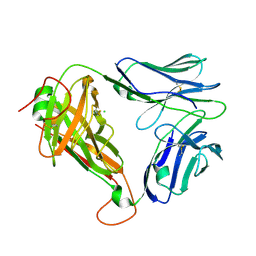 | | Crystal structure of parent MAGE-A10 TCR (728) | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T-cell receptor alpha chain (TRAV/TRAC), ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
7PDW
 
 | | Crystal structure of parent TCR (728) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
6TLB
 
 | | Plasmodium falciparum lipocalin (PF3D7_0925900) | | Descriptor: | GLYCEROL, SODIUM ION, Serine/threonine protein kinase | | Authors: | Burda, P.C, Crosskey, T.D, Lauk, K, Wilmanns, M, Gilberger, T.W. | | Deposit date: | 2019-12-02 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (2.85 Å), X-RAY DIFFRACTION | | Cite: | Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum.
Cell Rep, 31, 2020
|
|
6L1S
 
 | | Crystal structure of DUSP22 mutant_C88S | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Chang, C.C, Lyu, P.C. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3611 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6L9M
 
 | | H2-Ld complexed with AH1 peptide | | Descriptor: | H2-Ld, SER-PRO-SER-TYR-VAL-TYR-HIS-GLN-PHE, b2m | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4Z9B
 
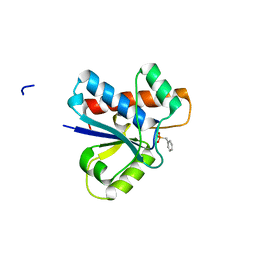 | | Crystal structure of Low Molecular Weight Protein Tyrosine Phosphatase isoform A complexed with benzylphosphonic acid | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase, benzylphosphonic acid | | Authors: | Fonseca, E.M.B, Trivella, D.B.B, Scorsato, V, Dias, M.P, de Oliveira, F.L, Miranda, P.C.M.L, Aparicio, R. | | Deposit date: | 2015-04-10 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of the apo form and a complex of human LMW-PTP with a phosphonic acid provide new evidence of a secondary site potentially related to the anchorage of natural substrates.
Bioorg.Med.Chem., 23, 2015
|
|
4ZWB
 
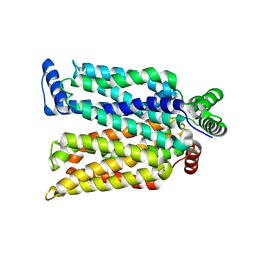 | | Crystal structure of maltose-bound human GLUT3 in the outward-occluded conformation at 2.4 angstrom | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 3, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
6LMY
 
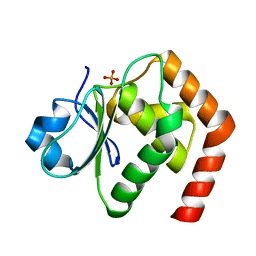 | | Crystal structure of DUSP22 mutant_C88S/S93A | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2019-12-27 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6LOT
 
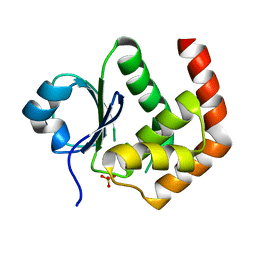 | | Crystal structure of DUSP22 mutant_N128D | | Descriptor: | Dual specificity protein phosphatase 22, SULFATE ION | | Authors: | Lai, C.H, Lyu, P.C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
6M4T
 
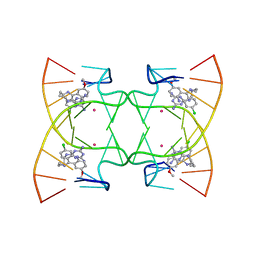 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*(UD)P*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), N4-[4-[(6-chloranyl-2-methoxy-acridin-9-yl)amino]butyl]-1,3,5-triazine-2,4,6-triamine | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B, Wu, P.C. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
4ZWC
 
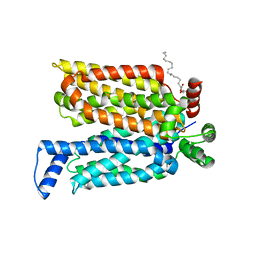 | | Crystal structure of maltose-bound human GLUT3 in the outward-open conformation at 2.6 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZW9
 
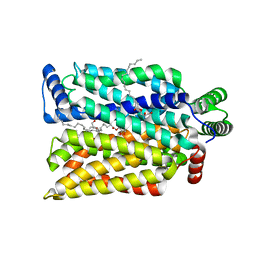 | | Crystal structure of human GLUT3 bound to D-glucose in the outward-occluded conformation at 1.5 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
5A0Q
 
 | |
7BI1
 
 | | XFEL crystal structure of soybean ascorbate peroxidase compound II | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kwon, H, Tosha, T, Sugimoto, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2021-01-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XFEL Crystal Structures of Peroxidase Compound II.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7BIU
 
 | | XFEL crystal structure of cytochrome c peroxidase compound II | | Descriptor: | Cytochrome c peroxidase, mitochondrial, HEME C | | Authors: | Kwon, H, Tosha, T, Sugimoto, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | XFEL Crystal Structures of Peroxidase Compound II.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B5J
 
 | | Anti-CRISPR associated (Aca) protein, Aca2 | | Descriptor: | Anti-CRISPR associated (Aca) protein, Aca2, GLYCEROL | | Authors: | Usher, B, Birkholz, N, Fineran, P.C, Blower, T.R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the anti-CRISPR repressor Aca2.
J.Struct.Biol., 213, 2021
|
|
7BEW
 
 | |
7BEX
 
 | |
8G6I
 
 | | Coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VIII chimera from human and pig, NB33 heavy chain, ... | | Authors: | Childers, K.C, Davulcu, O, Haynes, R.M, Lollar, P, Doering, C.B, Coxon, C.H, Spiegel, P.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor.
Blood, 142, 2023
|
|
2PQJ
 
 | | Crystal structure of active ribosome inactivating protein from maize (b-32), complex with adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Au, S.W.N, Cha, S.S, Young, J.A, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
2PMW
 
 | | The Crystal Structure of Proprotein convertase subtilisin kexin type 9 (PCSK9) | | Descriptor: | Proprotein convertase subtilisin/kexin type 9, SULFATE ION | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of PCSK9: A Regulator of Plasma LDL-Cholesterol.
Structure, 15, 2007
|
|
2PNI
 
 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
