1C2P
 
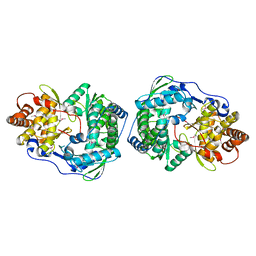 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Lesburg, C.A, Cable, M.B, Ferrari, E, Hong, Z, Mannarino, A.F, Weber, P.C. | | Deposit date: | 1999-07-26 | | Release date: | 2000-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site.
Nat.Struct.Biol., 6, 1999
|
|
5V53
 
 | |
3CPL
 
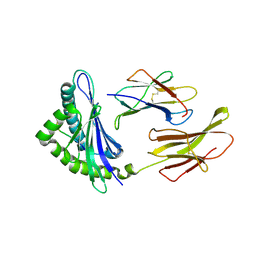 | | Crystal Structure of H-2Db in complex with a variant M6A of the NP366 peptide from influenza A virus | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Kedzierska, K, Guillonneau, C, Hatton, L.A, Stockwell, D, Gras, S, Webby, R, Rossjohn, J, Purcell, A.W, Doherty, P.C, Turner, S.J. | | Deposit date: | 2008-03-31 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complete modification of TCR specificity and repertoire selection does not perturb a CD8+ T cell immunodominance hierarchy.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5DAU
 
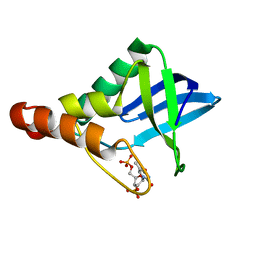 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39E/V104E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Robinson, A.C, Siegler, M.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-08-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39E/V104E at cryogenic temperature
To be Published
|
|
4O7J
 
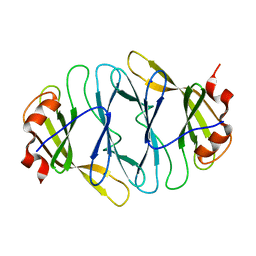 | |
1SIY
 
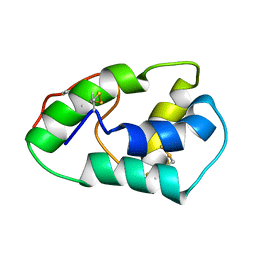 | | NMR structure of mung bean non-specific lipid transfer protein 1 | | Descriptor: | Nonspecific lipid-transfer protein 1 | | Authors: | Lin, K.F, Liu, Y.N, Hsu, S.T.D, Samuel, D, Cheng, C.S, Bonvin, A.M.J.J, Lyu, P.C. | | Deposit date: | 2004-03-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Characterization and Structural Analyses of Nonspecific Lipid Transfer Protein 1 from Mung Bean
Biochemistry, 44, 2005
|
|
8I1B
 
 | |
7P46
 
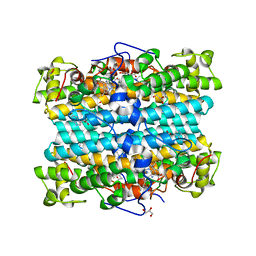 | | Crystal Structure of Xanthomonas campestris Tryptophan 2,3-dioxygenase (TDO) | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, CYANIDE ION, GLYCEROL, ... | | Authors: | Kwon, H, Basran, J, Booth, E.S, Campbell, L.P, Thackray, S.J, Moody, P.C.E, Mowat, C.G, Raven, E.L. | | Deposit date: | 2021-07-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of l-kynurenine to X. campestris tryptophan 2,3-dioxygenase.
J.Inorg.Biochem., 225, 2021
|
|
8PM2
 
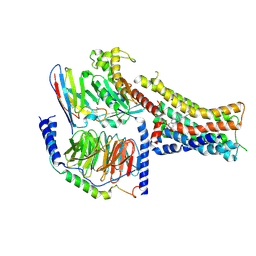 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
3UT2
 
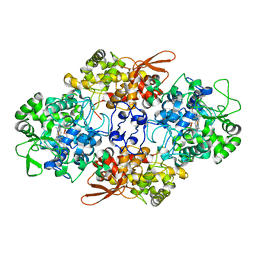 | | Crystal Structure of Fungal MagKatG2 | | Descriptor: | Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zamocky, M, Garcia-Fernandez, M.Q, Gasselhuber, B, Jakopitsch, C, Furtmuller, P.G, Loewen, P.C, Fita, I, Obinger, C, Carpena, X. | | Deposit date: | 2011-11-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | High Conformational Stability of Secreted Eukaryotic Catalase-peroxidases: ANSWERS FROM FIRST CRYSTAL STRUCTURE AND UNFOLDING STUDIES.
J.Biol.Chem., 287, 2012
|
|
8DT3
 
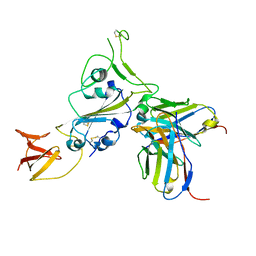 | | Cryo-EM structure of spike binding to Fab of neutralizing antibody (locally refined) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain Fab of SW186, Light chain Fab of SW186, ... | | Authors: | Sun, P.C, Fang, Y, Bai, X.C, Chen, Z.J. | | Deposit date: | 2022-07-25 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An antibody that neutralizes SARS-CoV-1 and SARS-CoV-2 by binding to a conserved spike epitope outside the receptor binding motif.
Sci Immunol, 7, 2022
|
|
7B5J
 
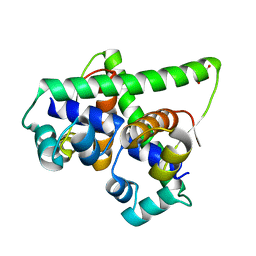 | | Anti-CRISPR associated (Aca) protein, Aca2 | | Descriptor: | Anti-CRISPR associated (Aca) protein, Aca2, GLYCEROL | | Authors: | Usher, B, Birkholz, N, Fineran, P.C, Blower, T.R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the anti-CRISPR repressor Aca2.
J.Struct.Biol., 213, 2021
|
|
1JP3
 
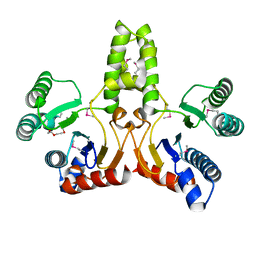 | | Structure of E.coli undecaprenyl pyrophosphate synthase | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, undecaprenyl pyrophosphate synthase | | Authors: | Ko, T.P, Chen, Y.K, Robinson, H, Tsai, P.C, Gao, Y.G, Chen, A.P.C, Wang, A.H.J, Liang, P.H. | | Deposit date: | 2001-07-31 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis.
J.Biol.Chem., 276, 2001
|
|
6JOF
 
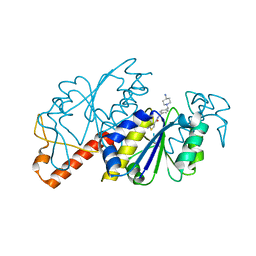 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with active-site inhibitor | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase, ~{N}-[[4-[(4-azanylpiperidin-1-yl)methyl]phenyl]methyl]-4-oxidanylidene-3~{H}-thieno[2,3-d]pyrimidine-5-carboxamide | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2019-03-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
6JKI
 
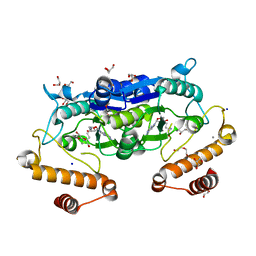 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
6WJG
 
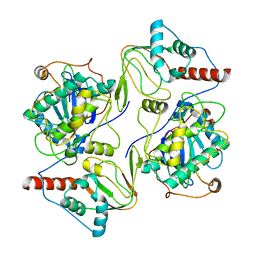 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
6WJF
 
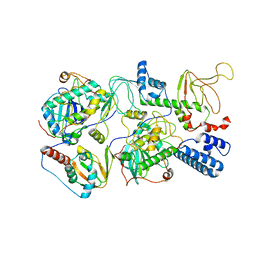 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
6TAE
 
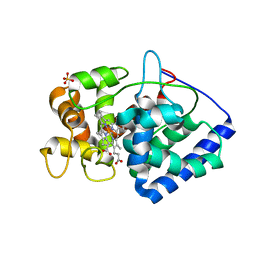 | | Neutron structure of ferric ascorbate peroxidase | | Descriptor: | Ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4G41
 
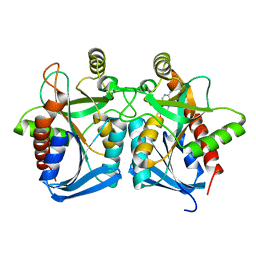 | | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Ponniah, K, Norris, G.E, Anderson, B.F, Brown, R.L, Tyler, P.C, Evans, G.B, Frohlich, R. | | Deposit date: | 2012-07-15 | | Release date: | 2012-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin;
TO BE PUBLISHED
|
|
7BI1
 
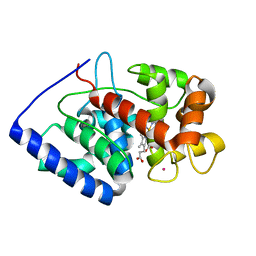 | | XFEL crystal structure of soybean ascorbate peroxidase compound II | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kwon, H, Tosha, T, Sugimoto, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2021-01-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XFEL Crystal Structures of Peroxidase Compound II.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7BIU
 
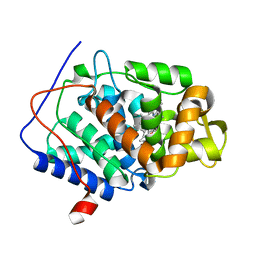 | | XFEL crystal structure of cytochrome c peroxidase compound II | | Descriptor: | Cytochrome c peroxidase, mitochondrial, HEME C | | Authors: | Kwon, H, Tosha, T, Sugimoto, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | XFEL Crystal Structures of Peroxidase Compound II.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4G89
 
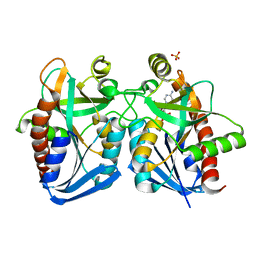 | | Crystal structure of k. pneumoniae mta/adohcy nucleosidase in complex with fragmented s-adenosyl-l-homocysteine | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Norris, G.E, Brown, R.L, Anderson, B.F, Tyler, P.C, Evans, G.B. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of k. pneumoniae mta/adohcy nucleosidase in complex with fragmented s-adenosyl-l-homocysteine
To be Published
|
|
6XV4
 
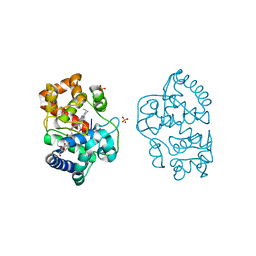 | | Neutron structure of ferric ascorbate peroxidase-ascorbate complex | | Descriptor: | ASCORBIC ACID, Ascorbate peroxidase, POTASSIUM ION, ... | | Authors: | Kwon, H, Basran, J, Devos, J.M, Schrader, T.E, Ostermann, A, Blakeley, M.P, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2020-01-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Visualizing the protons in a metalloenzyme electron proton transfer pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UNA
 
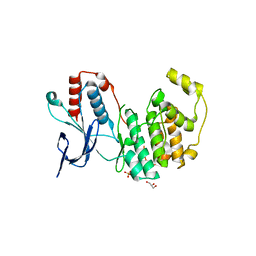 | | Crystal structure of inactive p38gamma | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 12, SULFATE ION | | Authors: | Aoto, P.C, Stanfield, R.L, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-10-11 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A Dynamic Switch in Inactive p38 gamma Leads to an Excited State on the Pathway to an Active Kinase.
Biochemistry, 58, 2019
|
|
6XLQ
 
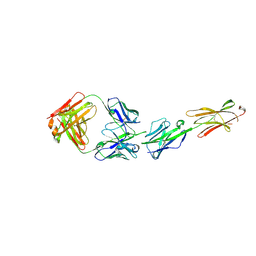 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
