3GG6
 
 | | Crystal structure of the NUDIX domain of human NUDT18 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 18 | | Authors: | Tresaugues, L, Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NUDIX domain of human NUDT18
To be Published
|
|
5PZ7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 103) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2V43
 
 | |
2OZF
 
 | | The crystal structure of the 2nd PDZ domain of the human NHERF-1 (SLC9A3R1) | | Descriptor: | Ezrin-radixin-moesin-binding phosphoprotein 50 | | Authors: | Phillips, C, Papagrigoriou, E, Gileadi, C, Fedorov, O, Elkins, J, Berridge, G, Turnbull, A.P, Gileadi, O, Schoch, G, Smee, C, Bray, J, Savitsky, P, Uppenberg, J, von Delft, F, Gorrec, F, Umeano, C, Salah, E, Colebrook, S, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 2nd PDZ domain of the human NHERF-1 (SLC9A3R1)
To be Published
|
|
1UW6
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
3GH3
 
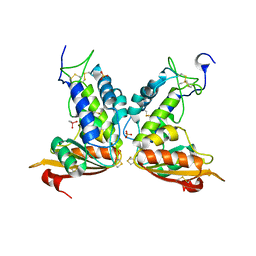 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, Ecto-NAD+ glycohydrolase (CD38 molecule), ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Kellenberger, E, Oppenheimer, N, Schuber, F. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
6HY7
 
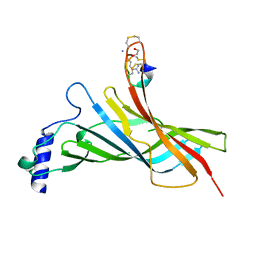 | |
5X45
 
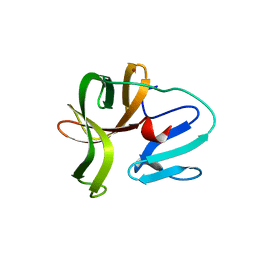 | | Crystal structure of 2A protease from Human rhinovirus C15 | | Descriptor: | ZINC ION, protease 2A | | Authors: | Ling, H, Yang, P, Shaw, N, Sun, Y, Wang, X. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural view of the 2A protease from human rhinovirus C15.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
1URL
 
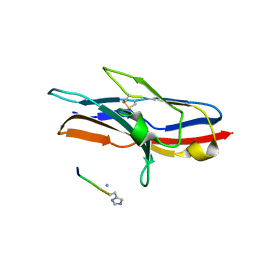 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH GLYCOPEPTIDE | | Descriptor: | ALA-GLY-HIS-THR-TRP-GLY-HIA, N-acetyl-alpha-neuraminic acid, SIALOADHESIN | | Authors: | Bukrinsky, J.T, Hilaire, P.M.S, Meldal, M, Crocker, P.R, Henriksen, A. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex of Sialoadhesin with a Glycopeptide Ligand
Biochim.Biophys.Acta, 1702, 2004
|
|
1V2I
 
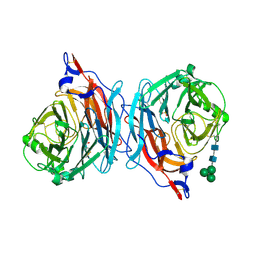 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1LBG
 
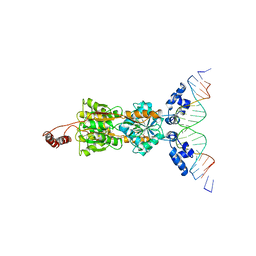 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
5PEM
 
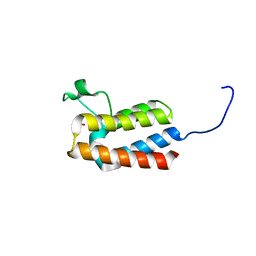 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 115) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1UTC
 
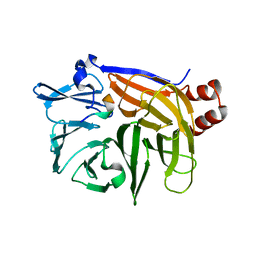 | | Clathrin terminal domain complexed with TLPWDLWTT | | Descriptor: | AMPHIPHYSIN, CLATHRIN HEAVY CHAIN | | Authors: | Miele, A.E, Evans, P.R, Owen, D.J. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller.
Nat. Struct. Mol. Biol., 11, 2004
|
|
4Q3N
 
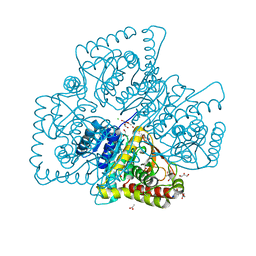 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
5PF6
 
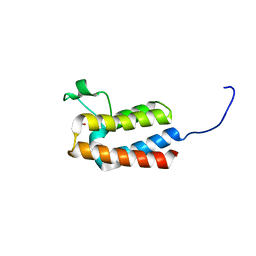 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 135) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2OUB
 
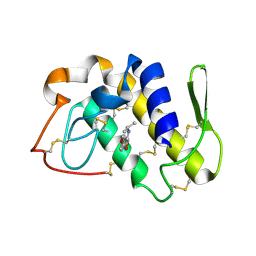 | | Crystal structure of the complex formed between phospholipase A2 and atenolol at 2.75 A resolution | | Descriptor: | 2-(4-(2-HYDROXY-3-(ISOPROPYLAMINO)PROPOXY)PHENYL)ETHANAMIDE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-10 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed between phospholipase A2 and atenolol at 2.75 A resolution
To be Published
|
|
5PFT
 
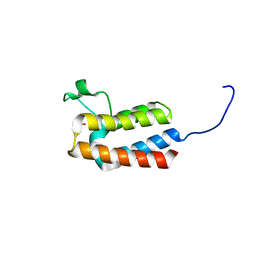 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 157) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4Q3G
 
 | | Structure of the OsSERK2 leucine rich repeat extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, OsSERK2 | | Authors: | McAndrew, R.P, Pruitt, R.N, Kamita, S.G, Pereira, J.H, Majumder, D, Hammock, B.D, Adams, P.D, Ronald, P.C. | | Deposit date: | 2014-04-11 | | Release date: | 2014-11-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Structure of the OsSERK2 leucine-rich repeat extracellular domain.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
5PG7
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 171) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PGR
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 190) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1AEW
 
 | | L-CHAIN HORSE APOFERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Hempstead, P.D, Yewdall, S.J, Lawson, D.M, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution.
J.Mol.Biol., 268, 1997
|
|
2UUH
 
 | | Crystal structure of Human Leukotriene C4 Synthase in complex with substrate glutathione | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, GLUTATHIONE, LEUKOTRIENE C4 SYNTHASE, ... | | Authors: | Martinez Molina, D, Wetterholm, A, Kohl, A, McCarthy, A.A, Niegowski, D, Ohlson, E, Hammarberg, T, Eshaghi, S, Haeggstrom, J.Z, Nordlund, P. | | Deposit date: | 2007-03-02 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Synthesis of Inflammatory Mediators by Human Leukotriene C4 Synthase.
Nature, 448, 2007
|
|
1LEJ
 
 | | NMR Structure of a 1:1 Complex of Polyamide (Im-Py-Beta-Im-Beta-Im-Py-Beta-Dp) with the Tridecamer DNA Duplex 5'-CCAAAGAGAAGCG-3' | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*AP*GP*AP*AP*GP*CP*G)-3', 5'-D(*CP*GP*CP*TP*TP*CP*TP*CP*TP*TP*TP*GP*G)-3', IMIDAZOLE-PYRROLE-BETA ALANINE-IMIDAZOLE-BETA ALANINE-IMIDAZOLE-PYRROLE-BETA ALANINE-DIMETHYLAMINO PROPYLAMIDE | | Authors: | Urbach, A.R, Love, J.J, Ross, S.A, Dervan, P.B. | | Deposit date: | 2002-04-09 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a beta-alanine-linked polyamide bound to a full helical turn of purine tract DNA in the 1:1 motif.
J.Mol.Biol., 320, 2002
|
|
2OW0
 
 | | MMP-9 active site mutant with iodine-labeled carboxylate inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix metalloproteinase-9 (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB), ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
