4BSI
 
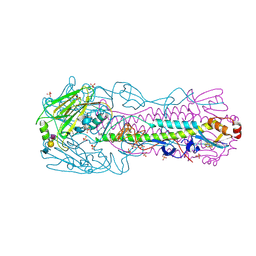 | | H7N3 Avian Influenza Virus Haemagglutinin in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Haire, L.F, Martin, S.R, Wharton, S.A, Daniels, R.S, Bennett, M.S, McCauley, J.W, Collins, P.J, Walker, P.A, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Receptor Binding by an H7N9 Influenza Virus from Humans
Nature, 499, 2013
|
|
8TM2
 
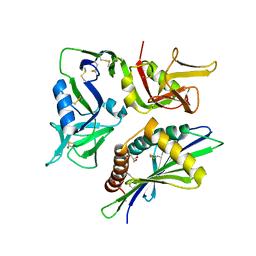 | |
6MZN
 
 | |
3COG
 
 | | Crystal structure of human cystathionase (Cystathionine gamma lyase) in complex with DL-propargylglycine | | Descriptor: | (2S)-2-aminopent-4-enoic acid, Cystathionine gamma-lyase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Collins, R, Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Schuler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Sagermark, J, Busam, R.D, Welin, M, Weigelt, J, Wikstrom, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S.
J.Biol.Chem., 284, 2009
|
|
4HCW
 
 | |
6W8S
 
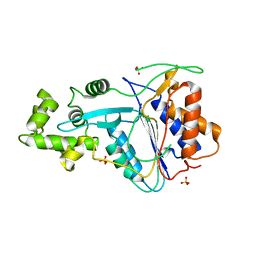 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
3AJA
 
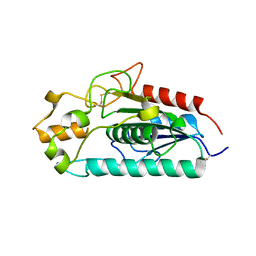 | |
6MR1
 
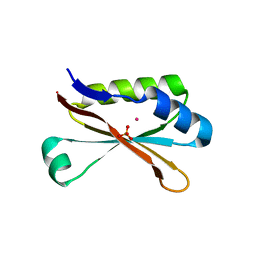 | | RbcS-like subdomain of CcmM | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Carbon dioxide concentrating mechanism protein, ... | | Authors: | Ryan, P, Kimber, M.S. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The small RbcS-like domains of the beta-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit.
J. Biol. Chem., 294, 2019
|
|
6VIP
 
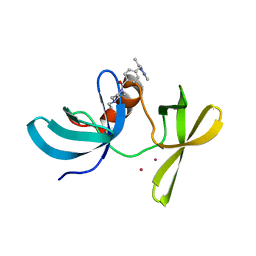 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008 | | Descriptor: | TP53-binding protein 1, UNKNOWN ATOM OR ION, {4-[(3,5-dimethyl-1H-pyrazol-1-yl)methyl]phenyl}(4-ethylpiperazin-1-yl)methanone | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008
to be published
|
|
3JBR
 
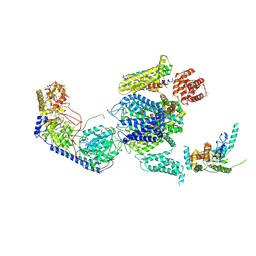 | | Cryo-EM structure of the rabbit voltage-gated calcium channel Cav1.1 complex at 4.2 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Voltage-dependent L-type calcium channel subunit alpha-1S, ... | | Authors: | Wu, J.P, Yan, Z, Yan, N. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 complex
Science, 350, 2015
|
|
1R42
 
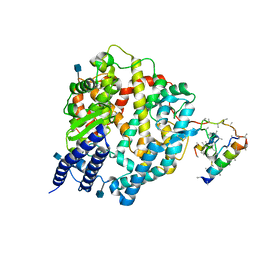 | | Native Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ZINC ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1R56
 
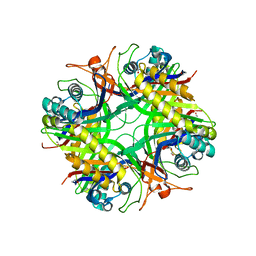 | | UNCOMPLEXED URATE OXIDASE FROM ASPERGILLUS FLAVUS | | Descriptor: | DI(HYDROXYETHYL)ETHER, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R70
 
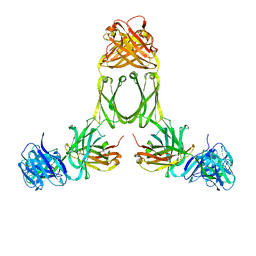 | | Model of human IgA2 determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Human IgA2(m1) Heavy Chain, Human IgA2(m1) Light Chain | | Authors: | Furtado, P.B, Whitty, P.W, Robertson, A, Eaton, J.T, Almogren, A, Kerr, M.A, Woof, J.M, Perkins, S.J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | Solution Structure Determination of Monomeric Human IgA2 by X-ray and Neutron Scattering, Analytical Ultracentrifugation and Constrained Modelling: A Comparison with Monomeric Human IgA1.
J.Mol.Biol., 338, 2004
|
|
4LJ2
 
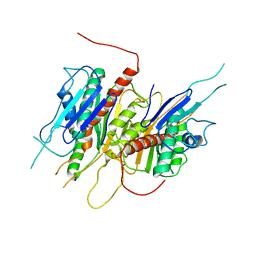 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 3.15A resolution | | Descriptor: | Chorismate synthase | | Authors: | Chaudhary, A, Singh, N, Kaushik, S, Tyagi, T.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-04 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 3.15A resolution
To be Published
|
|
3Q00
 
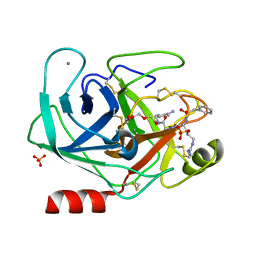 | | Bovine trypsin variant X(tripleGlu217Ile227) in complex with small molecule inhibitor | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-12-15 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4RHJ
 
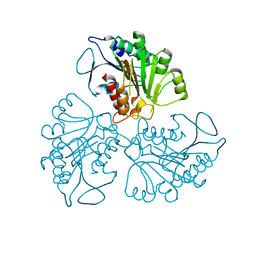 | |
6NVD
 
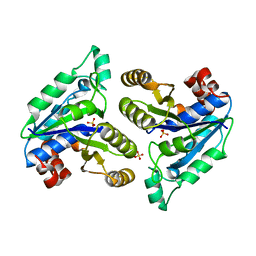 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9 | | Descriptor: | 2',5'-dideoxy-5'-[4-({[(1S,2R)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetyl}amino)-1H-1,2,3-triazol-1-yl]cytidine, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with triazole linked compound 9
To Be Published
|
|
5EAT
 
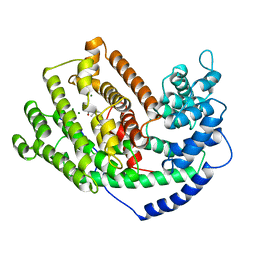 | | 5-EPI-ARISTOLOCHENE SYNTHASE FROM NICOTIANA TABACUM WITH SUBSTRATE ANALOG FARNESYL HYDROXYPHOSPHONATE | | Descriptor: | 1-HYDROXY-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENE PHOSPHONIC ACID, 5-EPI-ARISTOLOCHENE SYNTHASE, MAGNESIUM ION | | Authors: | Starks, C.M, Back, K, Chappell, J, Noel, J.P. | | Deposit date: | 1997-07-24 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase.
Science, 277, 1997
|
|
4CN2
 
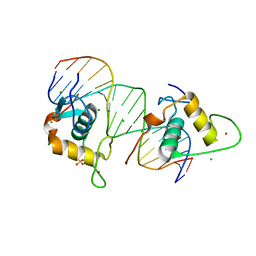 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Ramp2 Response Element | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*TP*TP*GP*AP*CP*CP*CP*TP*TP*GP*AP*AP*DC *TP*CP*AP)-3', 5'-D(*TP*GP*AP*GP*TP*TP*CP*AP*AP*GP*GP*GP*TP*DC *AP*AP*TP)-3', ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
4RJ2
 
 | |
3OA6
 
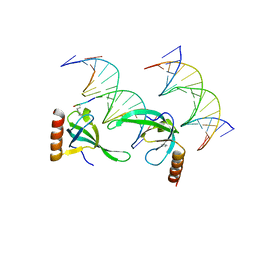 | | Human MSL3 Chromodomain bound to DNA and H4K20me1 peptide | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), H4 peptide monomethylated at lysine 20, ... | | Authors: | Kim, D, Huang, P, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2JH7
 
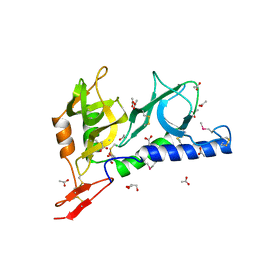 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 6'-sialyl-N-acetyllactosamine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
6NJ8
 
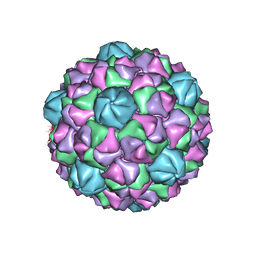 | | Encapsulin iron storage compartment from Quasibacillus thermotolerans | | Descriptor: | Encapsulating protein for a DyP-type peroxidase, targeting peptide | | Authors: | Orlando, B.J, Giessen, T.W, Chambers, M.G, Liao, M, Silver, P.A. | | Deposit date: | 2019-01-02 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Large protein organelles form a new iron sequestration system with high storage capacity.
Elife, 8, 2019
|
|
6G79
 
 | | Coupling specificity of heterotrimeric Go to the serotonin 5-HT1B receptor | | Descriptor: | 2-[5-[2-[4-(4-cyanophenyl)piperazin-1-yl]-2-oxidanylidene-ethoxy]-1~{H}-indol-3-yl]ethylazanium, 5-hydroxytryptamine receptor 1B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Garcia-Nafria, J, Nehme, R, Edwards, P, Tate, C.G. | | Deposit date: | 2018-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM structure of the serotonin 5-HT1Breceptor coupled to heterotrimeric Go.
Nature, 558, 2018
|
|
2JAK
 
 | | Human PP2A regulatory subunit B56g | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A 56 KDA REGULATORY SUBUNIT GAMMA ISOFORM | | Authors: | Magnusdottir, A, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Lundgren, S, Moche, M, Nilsson, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Upsten, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Pp2A Regulatory Subunit B56 Gamma: The Remaining Piece of the Pp2A Jigsaw Puzzle.
Proteins, 74, 2009
|
|
