4UT6
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 B7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 B7, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UTA
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 HEAVY CHAIN, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 LIGHT CHAIN, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
8Q2G
 
 | |
9FFB
 
 | | ss-dsDNA-FANCD2-FANCI complex | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*GP*TP*CP*TP*CP*TP*AP*GP*AP*CP*AP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*CP*TP*GP*TP*CP*TP*AP*GP*AP*GP*AP*CP*AP*TP*CP*GP*AP*T)-3'), Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Passmore, L.A. | | Deposit date: | 2024-05-22 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | ss-dsDNA-FANCD2-FANCI complex
To Be Published
|
|
9XIA
 
 | |
9ABP
 
 | |
4TN3
 
 | | Structure of the BBox-Coiled-coil region of Rhesus Trim5alpha | | Descriptor: | TRIM5/cyclophilin A fusion protein/T4 Lysozyme chimera, ZINC ION | | Authors: | Kirkpatrick, J.J, Stoye, J.P, Taylor, I.A, Goldstone, D.C. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1989 Å) | | Cite: | Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TR2
 
 | | Crystal structure of PvSUB1 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Subtilisin-like 1 serine protease | | Authors: | Giganti, D, Bouillon, A, Martinez, M, Weber, P, Girard-Blanc, C, Petres, S, Haouz, A, Barale, J.C, Alzari, P.M. | | Deposit date: | 2014-06-13 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel Plasmodium-specific prodomain fold regulates the malaria drug target SUB1 subtilase.
Nat Commun, 5, 2014
|
|
8ALX
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, AMINOMETHYLAMIDE, ACETATE ION, ... | | Authors: | Rodriguez, I, Grudnik, P, Holak, T, Magiera-Mularz, K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biological characterization of pAC65, a macrocyclic peptide that blocks PD-L1 with equivalent potency to the FDA-approved antibodies.
Mol Cancer, 22, 2023
|
|
4RWN
 
 | | Crystal structure of the pre-reactive state of porcine OAS1 | | Descriptor: | 2'-5'-oligoadenylate synthase 1, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8AY5
 
 | | Human rhinovirus 2 empty particle in situ | | Descriptor: | Capsid protein VP1, Capsid protein VP2, VP3 | | Authors: | Ishemgulova, A, Mukhamedova, L, Trebichalska, Z, Payne, P, Smerdova, L, Moravcova, J, Hrebik, D, Buchta, D, Skubnik, K, Fuzik, T, Novacek, J, Plevka, P. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Endosome rupture enables enteroviruses to infect cells.
To Be Published
|
|
8AY4
 
 | | Human rhinovirus 2 virion in situ | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Ishemgulova, A, Mukhamedova, L, Trebichalska, Z, Payne, P, Smerdova, L, Moravcova, J, Hrebik, D, Buchta, D, Skubnik, K, Fuzik, T, Novacek, J, Plevka, P. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Endosome rupture enables enteroviruses to infect cells.
To Be Published
|
|
7LHC
 
 | |
8XIA
 
 | |
4TT6
 
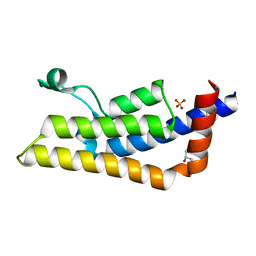 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
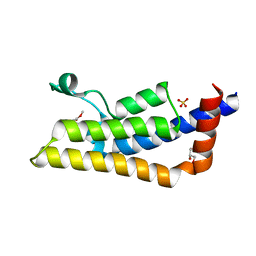 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
9BF6
 
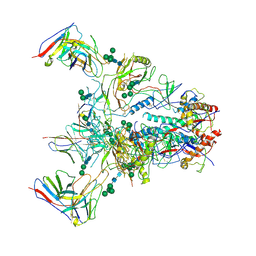 | |
4RWO
 
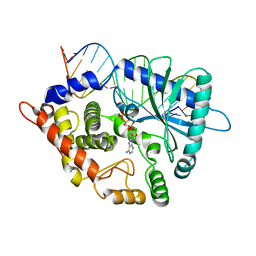 | | Crystal structure of the porcine OAS1 L149R mutant in complex with dsRNA and ApCpp in the AMP donor position | | Descriptor: | 2'-5'-oligoadenylate synthase 1, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
7MWA
 
 | | Crystal structure of 2-octaprenyl-6-methoxyphenol hydroxylase UbiH from Acinetobacter baumannii, apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-polyprenyl-6-methoxyphenol 4-hydroxylase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UbiH from Acinetobacter baumannii
To Be Published
|
|
4S2Q
 
 | | Crystal Structure of HMG domain of the chondrogenesis master regulator, Sox9 in complex with ChIP-Seq identified DNA element | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*GP*AP*AP*CP*AP*AP*AP*GP*CP*CP*TP*G)-3'), DNA (5'-D(P*AP*GP*GP*CP*TP*TP*TP*GP*TP*TP*CP*TP*CP*CP*TP*G)-3'), Transcription factor SOX-9 | | Authors: | Vivekanandan, S, Moovarkumudalvan, B, Lescar, J, Kolatkar, P.R. | | Deposit date: | 2015-01-21 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of HMG domain of the chondrogenesis master regulator, Sox9 in complex with ChIP-Seq identified DNA element
To be Published
|
|
8BW4
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-4-(3-fluoranylthiophen-2-yl)carbonyl-N-(4-methoxyphenyl)-2-methyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BAD
 
 | | Tpp80Aa1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Binary toxin A-like protein, CALCIUM ION, ... | | Authors: | Best, H.L, Williamson, L.J, Rizkallah, P.J, Berry, C, Lloyd-Evans, E. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure of Bacillus thuringiensis Tpp80Aa1 and Its Interaction with Galactose-Containing Glycolipids.
Toxins, 14, 2022
|
|
4TOA
 
 | | 1.95A resolution structure of Iron Bound BfrB (N148L) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
1ACB
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF THE BOVINE ALPHA-CHYMOTRYPSIN-EGLIN C COMPLEX AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-CHYMOTRYPSIN, Eglin C | | Authors: | Bolognesi, M, Frigerio, F, Coda, A, Pugliese, L, Lionetti, C, Menegatti, E, Amiconi, G, Schnebli, H.P, Ascenzi, P. | | Deposit date: | 1991-11-08 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and molecular structure of the bovine alpha-chymotrypsin-eglin c complex at 2.0 A resolution.
J.Mol.Biol., 225, 1992
|
|
