8D3Q
 
 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/NoPAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
7ASH
 
 | |
8D3M
 
 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/Processed prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
5FRM
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ384 (compound 4a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-azanylidene-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
7ZSA
 
 | |
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7ZS9
 
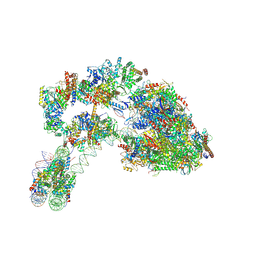 | |
8D3P
 
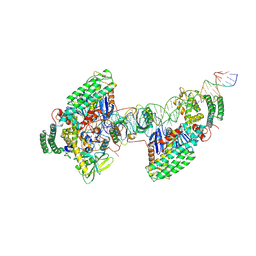 | | Type I-C Cas4-Cas1-Cas2 complex bound to half-site integration intermediate (HSI) | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
7Q0V
 
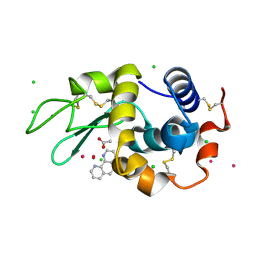 | | Lysozyme soaked with V(IV)OSO4 and phen | | Descriptor: | 1,10-PHENANTHROLINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Santos, M.F.A, Fernandes, A.C.P, Correia, I, Sciortino, G, Garribba, E, Santos-Silva, T, Pessoa, J.C. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Binding of V IV O 2+ , V IV OL, V IV OL 2 and V V O 2 L Moieties to Proteins: X-ray/Theoretical Characterization and Biological Implications.
Chemistry, 28, 2022
|
|
7N0M
 
 | | Inactivated state of 2-APB-bound wildtype rat TRPV2 in nanodiscs | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.A, Protopopova, A.D, Gallo, P.N, Moiseenkova-Bell, V.Y. | | Deposit date: | 2021-05-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into TRPV2 activation by small molecules.
Nat Commun, 13, 2022
|
|
7ZWD
 
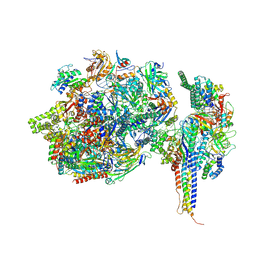 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DGA
 
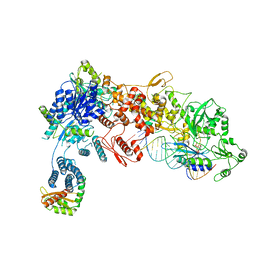 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IV | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8IQE
 
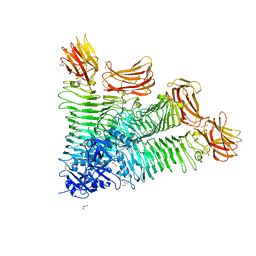 | | Crystal structure of tetrameric K2-2 TSP | | Descriptor: | GLYCEROL, K2-VCL6 TSP | | Authors: | Ye, T.J, Huang, K.F, Tu, I.F, Lee, I.M, Chang, Y.P, Wu, S.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
8FTC
 
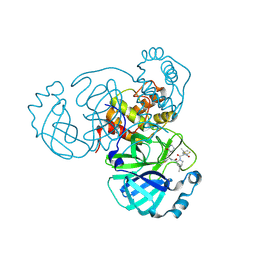 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | (1R,2S,5S)-3-[N-(difluoroacetyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopiperidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Chen, P, Khan, M.B, Lu, J, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Effect of Deuteration and Homologation of the Lactam Ring of Nirmatrelvir on Its Biochemical Properties and Oxidative Metabolism.
Acs Bio Med Chem Au, 3, 2023
|
|
7N0N
 
 | |
7ZX7
 
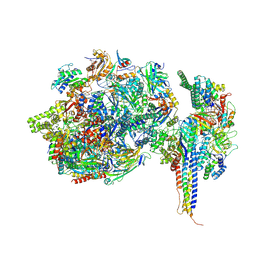 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6EZJ
 
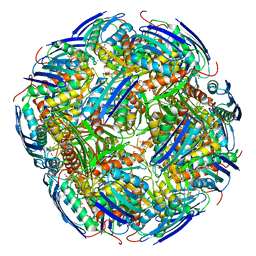 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8DG7
 
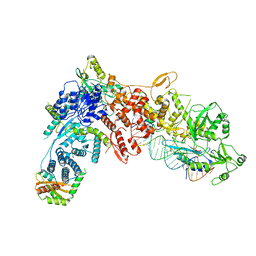 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex III | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8FWM
 
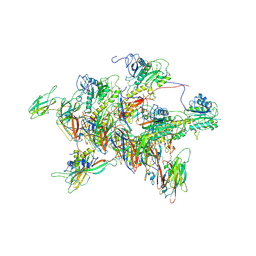 | | Structure of tail-neck junction of Agrobacterium phage Milano | | Descriptor: | Collar sheath protein, gp13, Tail sheath protein, ... | | Authors: | Sonani, R.R, Wang, F, Esteves, N.C, Kelly, R.J, Sebastian, A, Kreutzberger, M.A.B, Leiman, P.G, Scharf, B.E, Egelman, E.H. | | Deposit date: | 2023-01-23 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Neck and capsid architecture of the robust Agrobacterium phage Milano.
Commun Biol, 6, 2023
|
|
8DG5
 
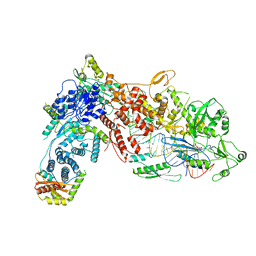 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
7ZHR
 
 | |
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3IB2
 
 | | structure of the complex of C-terminal half (C-lobe) of bovine lactoferrin with alpha-methyl-4-(2-methylpropyl) benzene acetic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Vikram, G, Kumar, R.P, Sinha, M, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2009-07-15 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The structural basis for the prevention of nonsteroidal antiinflammatory drug-induced gastrointestinal tract damage by the C-lobe of bovine colostrum lactoferrin.
Biophys.J., 97, 2009
|
|
8I8I
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Klebsiella pneumoniae at 2.59 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Phosphopantetheine adenylyltransferase | | Authors: | Ahmad, N, Singh, P.K, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of a novel form of phosphopantetheine adenylyltransferase from Klebsiella pneumoniae at 2.59 angstrom resolution.
Eur.Biophys.J., 53, 2024
|
|
7NMD
 
 | |
