6DC1
 
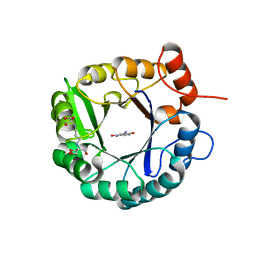 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 25 round 7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-nitro-2-oxidanyl-benzenecarbonitrile, ... | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
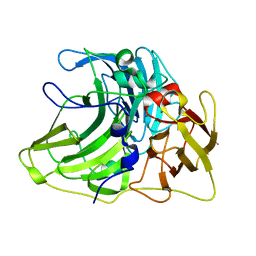
6Z0J
 
 | |
4EA9
 
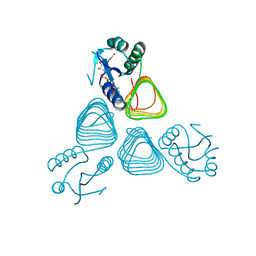 | | X-ray structure of GDP-perosamine N-acetyltransferase in complex with transition state analog at 0.9 Angstrom resolution | | Descriptor: | CHLORIDE ION, GDP-N-acetylperosamine-coenzyme A, Perosamine N-acetyltransferase | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4NGS
 
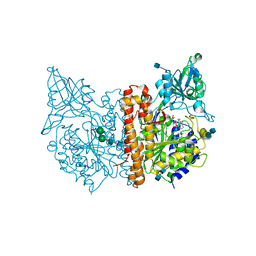 | | Crystal Structure of Glutamate Carboxypeptidase II in a complex with urea-based inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Pachl, P. | | Deposit date: | 2013-11-02 | | Release date: | 2014-06-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Rational design of urea-based glutamate carboxypeptidase II (GCPII) inhibitors as versatile tools for specific drug targeting and delivery.
Bioorg.Med.Chem., 22, 2014
|
|
5VYA
 
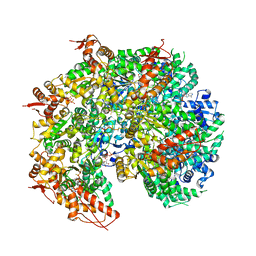 | | S. cerevisiae Hsp104:casein complex, Extended Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
6YUR
 
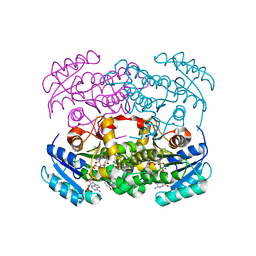 | | Crystal structure of S. aureus FabI inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Weinrich, J.D, Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
4ECH
 
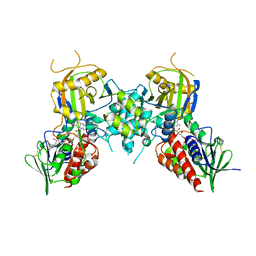 | | Yeast Polyamine Oxidase FMS1, H67Q Mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic and structural analyses of the role of his67 in the yeast polyamine oxidase fms1.
Biochemistry, 51, 2012
|
|
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | Descriptor: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
6Z0L
 
 | | Het-N2 - De novo designed three-helix heterodimer with Cysteine at the N2 position of the alpha-helix | | Descriptor: | CADMIUM ION, Cys-N2 Strand, Positive Strand, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7RPH
 
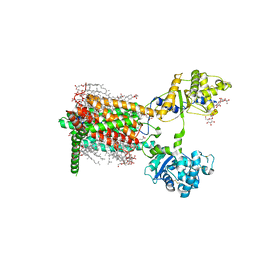 | | Cryo-EM structure of murine Dispatched 'R' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
6ZBO
 
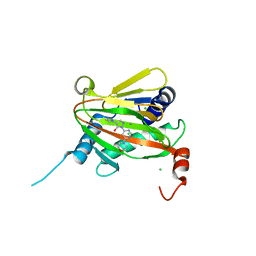 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in Complex with 1-(6-morpholinopyrimidin-4-yl)-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-5-ol (Molidustat) | | Descriptor: | 2-(6-morpholin-4-ylpyrimidin-4-yl)-4-(1,2,3-triazol-1-yl)pyrazol-3-ol, CHLORIDE ION, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
5VSA
 
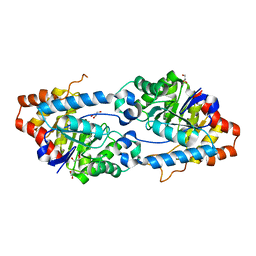 | | Crystal structure of SsoPox AsA1 mutant (C258L-I261F-W263A) | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-10 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
6Z4N
 
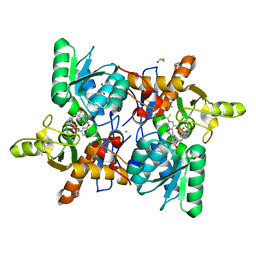 | | CRYSTAL STRUCTURE OF OASS COMPLEXED WITH UPAR INHIBITOR | | Descriptor: | (1~{S},2~{S})-1-[(4-methylphenyl)methyl]-2-phenyl-cyclopropane-1-carboxylic acid, COBALT (II) ION, Cysteine synthase A, ... | | Authors: | Demitri, N, Storici, P, Campanini, B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Investigational Studies on a Hit Compound Cyclopropane-Carboxylic Acid Derivative Targeting O -Acetylserine Sulfhydrylase as a Colistin Adjuvant.
Acs Infect Dis., 7, 2021
|
|
8AI6
 
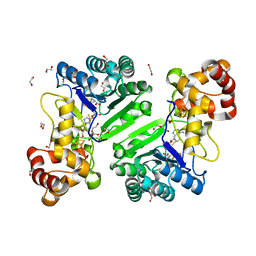 | | Crystal structure of radical SAM epimerase EpeE D210A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and persulfurated cysteine bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI1
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI4
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI3
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-methionine bound | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Chavas, L.M.G, Legrand, P, Polsinelli, I, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
1G8J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
8AI2
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI5
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
3TBS
 
 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX THE WITH LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (V3P,Y4A) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCOPROTEIN G1, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
5VW7
 
 | | NADPH soak of Y316A mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | ACETATE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
7RKH
 
 | | Yeast CTP Synthase (URA8) tetramer bound to ATP/UTP at neutral pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-22 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
4NGM
 
 | | Crystal Structure of Glutamate Carboxypeptidase II in a complex with urea-based inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Pachl, P. | | Deposit date: | 2013-11-02 | | Release date: | 2014-06-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Rational design of urea-based glutamate carboxypeptidase II (GCPII) inhibitors as versatile tools for specific drug targeting and delivery.
Bioorg.Med.Chem., 22, 2014
|
|
5W4T
 
 | | Crystal Structure of Fish Cadherin-23 EC1-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
