6Z7A
 
 | | Variant Surface Glycoprotein VSGsur | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E, Hashemi, H. | | Deposit date: | 2020-05-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | A Parasite Coat Protein Binds Suramin to Confer Drug Resistance
Nat Microbiol, 6, 2021
|
|
8B5Q
 
 | |
6WRQ
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) S158C variant bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
4WHG
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 3 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)octan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
7R0H
 
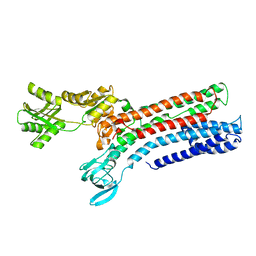 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | COPPER (II) ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0I
 
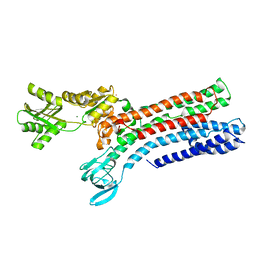 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
6QZP
 
 | | High-resolution cryo-EM structure of the human 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S rRNA (1740-MER), 28S rRNA (3773-MER), ... | | Authors: | Natchiar, S.K, Myasnikov, A.G, Kratzat, H, Hazemann, I, Klaholz, B.P. | | Deposit date: | 2019-03-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualization of chemical modifications in the human 80S ribosome structure.
Nature, 551, 2017
|
|
7R0G
 
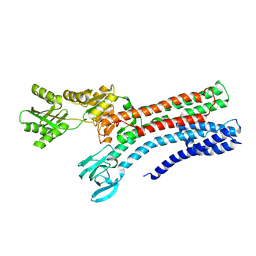 | |
8OET
 
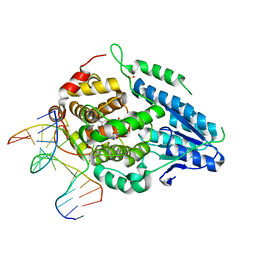 | | SFX structure of the class II photolyase complexed with a thymine dimer | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-03-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
7MM0
 
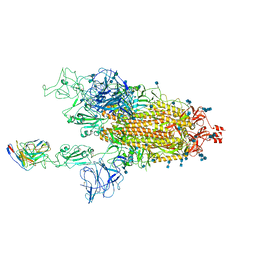 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
6OGZ
 
 | | In situ structure of Rotavirus RNA-dependent RNA polymerase at transcript-elongated state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Inner capsid protein VP2, RNA (5'-R(P*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*A)-3'), ... | | Authors: | Ding, K, Chang, T, Shen, W, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release.
Nat Commun, 10, 2019
|
|
6ZG9
 
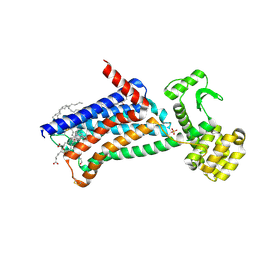 | | Structure of M1-StaR-T4L in complex with GSK1034702 at 2.5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 7-fluoranyl-5-methyl-3-[1-(oxan-4-yl)piperidin-4-yl]-1~{H}-benzimidazol-2-one, Muscarinic acetylcholine receptor M1,Endolysin,Muscarinic acetylcholine receptor M1, ... | | Authors: | Rucktooa, P, Cooke, R.M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From structure to clinic: Design of a muscarinic M1 receptor agonist with potential to treatment of Alzheimer's disease.
Cell, 184, 2021
|
|
8DTI
 
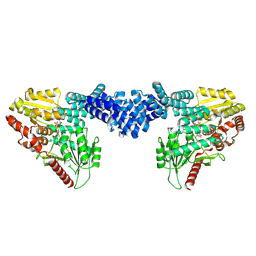 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
4WHF
 
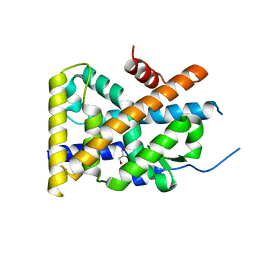 | | Crystal Structure of TR3 LBD in complex with 1-(3,4,5-trihydroxyphenyl)decan-1-one | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
8DTG
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
6OMB
 
 | | Cdc48 Hexamer (Subunits A to E) with substrate bound to the central pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
8DTH
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 2 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTF
 
 | | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
6O8C
 
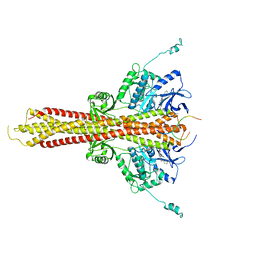 | | Crystal structure of STING CTT in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
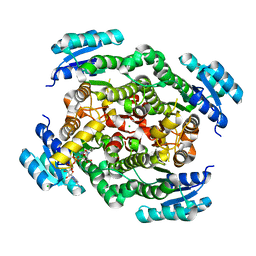
6ZZ0
 
 | |
6ZZT
 
 | |
6OPJ
 
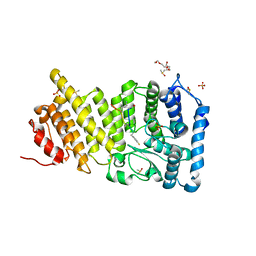 | | Menin in complex with peptide inhibitor 25 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, Peptide inhibitor 25, ... | | Authors: | Linhares, B.M, Fortuna, P, Cierpicki, T, Grembecka, J, Berlicki, L. | | Deposit date: | 2019-04-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5006572 Å) | | Cite: | Covalent and noncovalent constraints yield a figure eight-like conformation of a peptide inhibiting the menin-MLL interaction.
Eur.J.Med.Chem., 207, 2020
|
|
7ADV
 
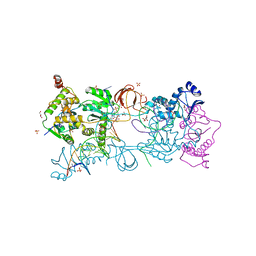 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ447 (compound 6v) | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[2-(2-morpholin-4-ylethylsulfonyl)ethyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Pye, V.E, Cherepanov, P. | | Deposit date: | 2020-09-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants.
Acs Infect Dis., 7, 2021
|
|
7ACK
 
 | | CDK2/cyclin A2 in complex with an imidazo[1,2-c]pyrimidin-5-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclohexyl-6~{H}-imidazo[1,2-c]pyrimidin-5-one, Cyclin-A2, ... | | Authors: | Skerlova, J, Pachl, P, Rezacova, P. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Imidazo[1,2-c]pyrimidin-5(6H)-one inhibitors of CDK2: Synthesis, kinase inhibition and co-crystal structure.
Eur.J.Med.Chem., 216, 2021
|
|
6OGY
 
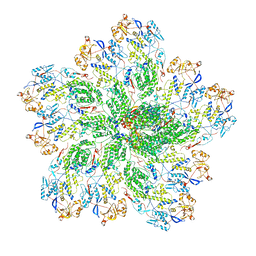 | | In situ structure of Rotavirus RNA-dependent RNA polymerase at duplex-open state | | Descriptor: | DNA/RNA (5'-D(*(GTG))-R(P*GP*C)-3'), Inner capsid protein VP2, RNA (5'-R(P*AP*GP*CP*C)-3'), ... | | Authors: | Ding, K, Chang, T, Shen, W, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release.
Nat Commun, 10, 2019
|
|
