3P6Z
 
 | | Structural basis of thrombin mediated factor V activation: essential role of the hirudin-like sequence Glu666-Glu672 for processing at the heavy chain-B domain junction | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corral-Rodriguez, M.A, Bock, P.E, Hernandez-Carvajal, E, Gutierrez-Gallego, R, Fuentes-Prior, P. | | Deposit date: | 2010-10-11 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of thrombin-mediated factor V activation: the Glu666-Glu672 sequence is critical for processing at the heavy chain-B domain junction.
Blood, 117, 2011
|
|
5FTT
 
 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
7U1Z
 
 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
5C0J
 
 | | HLA-A02 carrying RQFGPDWIVA | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5C0S
 
 | | Crystal structure of a generation 4 influenza hemagglutinin stabilized stem in complex with the broadly neutralizing antibody CR6261 | | Descriptor: | CR6261 antibody heavy chain, CR6261 antibody light chain, Hemagglutinin, ... | | Authors: | Boyington, J.C, Kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
5WBY
 
 | |
7TYY
 
 | | Human Amylin2 Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
3OS9
 
 | | Estrogen Receptor | | Descriptor: | 4-[1-allyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
6FM9
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
2XDB
 
 | | A processed non-coding RNA regulates a bacterial antiviral system | | Descriptor: | COBALT (II) ION, SULFATE ION, TOXI, ... | | Authors: | Blower, T.R, Pei, X.Y, Short, F.L, Fineran, P.C, Humphreys, D.P, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2010-04-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Processed Noncoding RNA Regulates an Altruistic Bacterial Antiviral System.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3OYH
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI MK0536 | | Descriptor: | 6-(3-chloro-4-fluorobenzyl)-4-hydroxy-N,N-dimethyl-2-(1-methylethyl)-3,5-dioxo-2,3,5,6,7,8-hexahydro-2,6-naphthyridine-1-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5T6W
 
 | | HLA-B*57:01 presenting SSTRGISQLW | | Descriptor: | Beta-2-microglobulin, Decapeptide: SER-SER-THR-ARG-GLY-ILE-SER-GLN-LEU-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5G0Z
 
 | | Structure of native granulovirus polyhedrin determined using an X-ray free-electron laser | | Descriptor: | GRANULIN | | Authors: | Gati, C, Bunker, R.D, Oberthur, D, Metcalf, P, Henry, C. | | Deposit date: | 2016-03-23 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2XB6
 
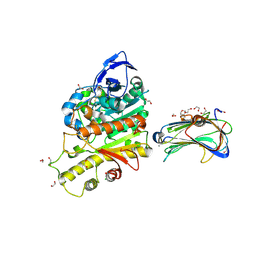 | | Revisited crystal structure of Neurexin1beta-Neuroligin4 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Leone, P, Comoletti, D, Ferracci, G, Conrod, S, Garcia, S.U, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into the Exquisite Selectivity of Neurexin-Neuroligin Synaptic Interactions
Embo J., 29, 2010
|
|
5THG
 
 | |
4EIX
 
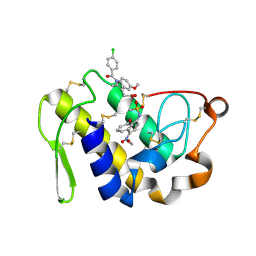 | | Structural Studies of the ternary complex of Phaspholipase A2 with nimesulide and indomethacin | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ACETONITRILE, INDOMETHACIN, ... | | Authors: | Shukla, P.K, Singh, N, Kumar, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Studies of the ternary complex of Phaspholipase A2 with nimusulide and indomethacin
To be Published
|
|
7TYX
 
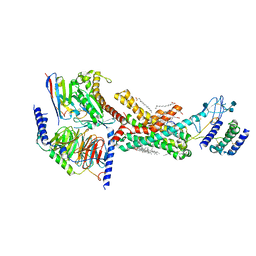 | | Human Amylin2 Receptor in complex with Gs and rat amylin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
3OYM
 
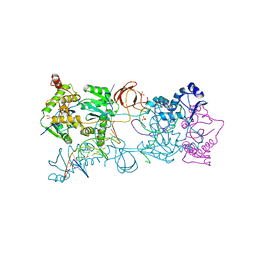 | | Crystal structure of the PFV N224H mutant intasome bound to manganese | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1GWC
 
 | | The structure of a tau class glutathione S-transferase from wheat, active in herbicide detoxification | | Descriptor: | GLUTATHIONE S-TRANSFERASE TSI-1, S-HEXYLGLUTATHIONE, SULFATE ION | | Authors: | Thom, R, Cummins, I, Dixon, D.P, Edwards, R, Cole, D.J, Lapthorn, A.J. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a Tau Class Glutathione S-Transferase from Wheat Active in Herbicide Detoxification
Biochemistry, 41, 2002
|
|
4R8J
 
 | | d(TCGGCGCCGA) with lambda-[Ru(TAP)2(dppz)]2+ soaked in D2O | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*(THM)P*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Hall, J.P, Gurung, S.P, Winter, G.W, Cardin, C.J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Monitoring one-electron photo-oxidation of guanine in DNA crystals using ultrafast infrared spectroscopy.
Nat Chem, 7, 2015
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
3LLN
 
 | | Comparison between the orthorhombic an tetragonal form of the heptamer sequence d(GCG(xT)GCG)/d(CGCACGC). | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*CP*GP*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*(XTR)P*GP*CP*G)-3') | | Authors: | Robeyns, K, Herdewijn, P, Van Meervelt, L. | | Deposit date: | 2010-01-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison between the orthorhombic and tetragonal forms of the heptamer sequence d[GCG(xT)GCG]/d(CGCACGC)
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5JUB
 
 | | Crystal structure of ComR from S.thermophilus in complex with DNA and its signalling peptide ComS. | | Descriptor: | ComS, Transcriptional regulator, pComX-for, ... | | Authors: | Talagas, A, Fontaine, L, Ledesma-Garcia, L, Li de la Sierra-Gallay, I, Hols, P, Nessler, S. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights into Streptococcal Competence Regulation by the Cell-to-Cell Communication System ComRS.
PLoS Pathog., 12, 2016
|
|
4ZA7
 
 | | Structure of A. niger Fdc1 in complex with alpha-methyl cinnamic acid | | Descriptor: | (2E)-2-methyl-3-phenylprop-2-enoic acid, 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
5T6G
 
 | | 2.45 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7m (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-(octylsulfonyl)-L-alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
