7PHO
 
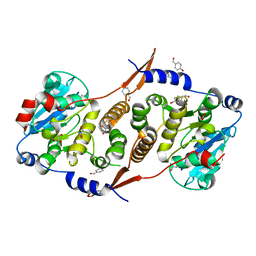 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde | | Descriptor: | BICARBONATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Nunes-Costa, D, Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-08-17 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde
To Be Published
|
|
4R15
 
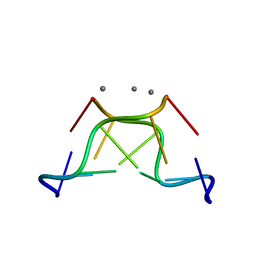 | | High-resolution crystal structure of Z-DNA in complex with Cr3+ cations | | Descriptor: | CHROMIUM ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2014-08-04 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations.
J.Biol.Inorg.Chem., 20, 2015
|
|
7O7O
 
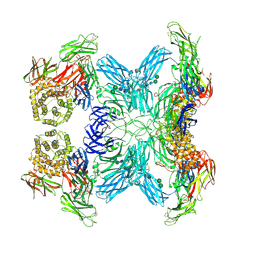 | | (h-alpha2M)4 semiactivated II state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8Q6B
 
 | | The RSL-D32N - sulfonato-calix[8]arene complex, I23 form, citrate pH 4.0, obtained by cross-seeding | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
8TH7
 
 | |
8D5H
 
 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin
To Be Published
|
|
156D
 
 | |
4R7N
 
 | | Fab C2E3 | | Descriptor: | Fab C2E3 Heavy chain, Fab C2E3 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-28 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
8TZZ
 
 | | SpG Cas9 with NGC PAM DNA target | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | Deposit date: | 2023-08-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8U3Y
 
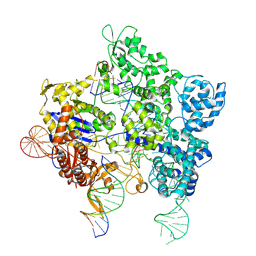 | | SpG Cas9 with NGG PAM DNA target | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8TH6
 
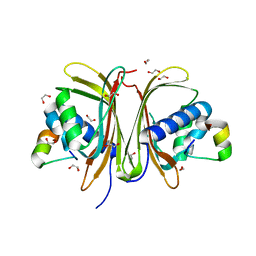 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Hughes, M.P, Taylor, J.P, Yang, Z. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
5K47
 
 | | CryoEM structure of the human Polycystin-2/PKD2 TRP channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Pike, A.C.W, Grieben, M, Shintre, C.A, Tessitore, A, Shrestha, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-24 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2).
Nat. Struct. Mol. Biol., 24, 2017
|
|
6G15
 
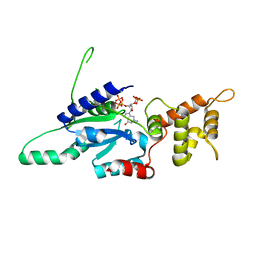 | | Crystal structure of pppGpp bound RbgA from S. aureus | | Descriptor: | Ribosome biogenesis GTPase A, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
5NAB
 
 | |
5KAQ
 
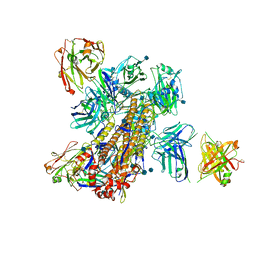 | | Crystal structure of broadly neutralizing Influenza A antibody 31.a.83 in complex with Hemagglutinin Hong Kong 1968. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 31.A.83 FAB HEAVY CHAIN, ANTIBODY 31.A.83 FAB LIGHT CHAIN, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
1AIL
 
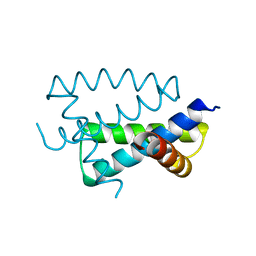 | | N-TERMINAL FRAGMENT OF NS1 PROTEIN FROM INFLUENZA A VIRUS | | Descriptor: | NONSTRUCTURAL PROTEIN NS1 | | Authors: | Liu, J, Lynch, P.A, Chien, C, Montelione, G.T, Krug, R.M, Berman, H.M. | | Deposit date: | 1997-04-21 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein.
Nat.Struct.Biol., 4, 1997
|
|
5K5B
 
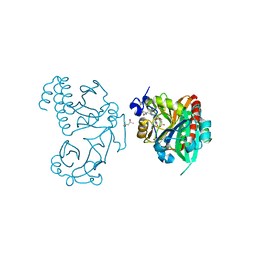 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans BphP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, ... | | Authors: | Takala, H, Edlund, P, Claesson, E, Ihalainen, J.A, Westenhoff, S. | | Deposit date: | 2016-05-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
6G7K
 
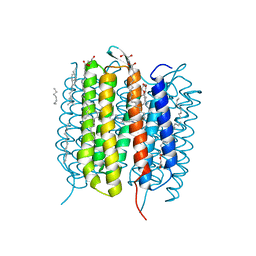 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 10 ps state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
5H0T
 
 | | Crystal structure of proliferating cell nuclear antigen from Leishmania donovani at 2.73 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Proliferating cell nuclear antigen | | Authors: | Singh, P.K, Yadav, S.P, Sharma, P.S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and binding studies of proliferating cell nuclear antigen from Leishmania donovani.
Biochim. Biophys. Acta, 1865, 2017
|
|
5KGN
 
 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
7EZJ
 
 | | Crystal structure of p73 DNA binding domain complex bound with 1 bp and 2 bp spacer DNA response elements. | | Descriptor: | 12-mer DNA, Tumor protein p73, ZINC ION | | Authors: | Koley, T, Roy Chowdhury, S, Kumar, M, Kaur, P, Singh, T.P, Viadiu, H, Ethayathulla, A.S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Deciphering the mechanism of p73 recognition of p53 response elements using the crystal structure of p73-DNA complexes and computational studies.
Int.J.Biol.Macromol., 206, 2022
|
|
8DIT
 
 | | Cryo-EM structure of a HOPS core complex containing Vps33, Vps16, and Vps18 | | Descriptor: | Vacuolar protein sorting-associated protein 16, Vacuolar protein sorting-associated protein 18, Vacuolar protein sorting-associated protein 33 | | Authors: | Port, S.A, Farrell, P.D, Jeffrey, P.D, DiMaio, F, Hughson, F.M. | | Deposit date: | 2022-06-29 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM structure of the HOPS core complex and its implication for SNARE assembly
To Be Published
|
|
3CWH
 
 | | D-xylose Isomerase in complex with linear product, per-deuterated xylulose | | Descriptor: | D-XYLULOSE, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P, Glusker, J.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Hydrogen location in stages of an enzyme-catalyzed reaction: time-of-flight neutron structure of D-xylose isomerase with bound D-xylulose
Biochemistry, 47, 2008
|
|
7NUP
 
 | | Endoplasmic reticulum aminopeptidase 2 complexed with a mixed hydroxamic and sulfonyl ligand | | Descriptor: | (3~{S})-4-(4-hydroxyphenyl)-~{N}-oxidanyl-3-[5-[[(5-phenylthiophen-2-yl)sulfonylamino]methyl]-1,2,3-triazol-1-yl]butanamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Stratikos, E. | | Deposit date: | 2021-03-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Endoplasmic reticulum aminopeptidase 2 complexed with a mixed hydroxamic and sulfonyl ligand
To Be Published
|
|
