6O26
 
 | | Crystal structure of 3246 Fab in complex with circumsporozoite protein NANA | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3246 Fab heavy chain, ... | | 著者 | Scally, S.W, Bosch, A, Castro, K, Murugan, R, Wardemann, H, Julien, J.P. | | 登録日 | 2019-02-22 | | 公開日 | 2020-03-04 | | 最終更新日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
6O2H
 
 | |
8DQJ
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (AST) bound to ATP and acridone | | 分子名称: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE MONOPHOSPHATE, ... | | 著者 | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | 登録日 | 2022-07-19 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
6W8R
 
 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | 分子名称: | Metacaspase-4, SULFATE ION | | 著者 | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | 登録日 | 2020-03-21 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6H0C
 
 | | Flv1 flavodiiron core from Synechocystis sp. PCC6803 | | 分子名称: | CHLORIDE ION, CITRATE ANION, Putative diflavin flavoprotein A 3 | | 著者 | Borges, P.T, Romao, C.V, Saraiva, L, Goncalves, V.L, Carrondo, M.A, Teixeira, M, Frazao, C. | | 登録日 | 2018-07-08 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.592 Å) | | 主引用文献 | Analysis of a new flavodiiron core structural arrangement in Flv1-Delta FlR protein from Synechocystis sp. PCC6803.
J. Struct. Biol., 205, 2019
|
|
6VRY
 
 | |
1T6H
 
 | | Crystal Structure T4 Lysozyme incorporating an unnatural amino acid p-iodo-L-phenylalanine at position 153 | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Spraggon, G, Xie, J, Wang, L, Wu, N, Brock, A, Schultz, P.G. | | 登録日 | 2004-05-06 | | 公開日 | 2004-10-26 | | 最終更新日 | 2018-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination.
Nat.Biotechnol., 22, 2004
|
|
3MLT
 
 | |
6VX5
 
 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | 分子名称: | Bestrophin, CHLORIDE ION | | 著者 | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | 登録日 | 2020-02-21 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2ZJQ
 
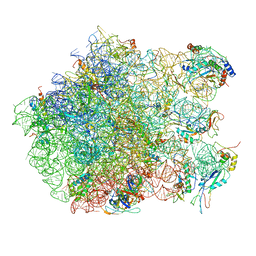 | | Interaction of L7 with L11 induced by Microccocin binding to the Deinococcus radiodurans 50S subunit | | 分子名称: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | 登録日 | 2008-03-08 | | 公開日 | 2008-06-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin.
Mol.Cell, 30, 2008
|
|
3FOH
 
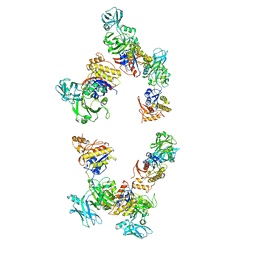 | | Fitting of gp18M crystal structure into 3D cryo-EM reconstruction of bacteriophage T4 extended tail | | 分子名称: | Tail sheath protein Gp18 | | 著者 | Aksyuk, A.A, Leiman, P.G, Kurochkina, L.P, Shneider, M.M, Kostyuchenko, V.A, Mesyanzhinov, V.V, Rossmann, M.G. | | 登録日 | 2008-12-30 | | 公開日 | 2009-03-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria.
Embo J., 28, 2009
|
|
6VX6
 
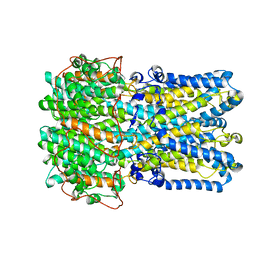 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | 分子名称: | Bestrophin, CALCIUM ION, CHLORIDE ION | | 著者 | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | 登録日 | 2020-02-21 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6NTP
 
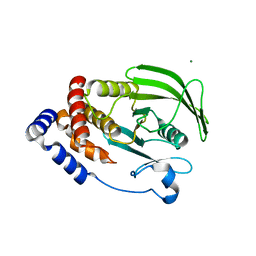 | | PTP1B Domain of PTP1B-LOV2 Chimera | | 分子名称: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1,NPH1-1 | | 著者 | Hongdusit, A, Sankaran, B, Zwart, P.H, Fox, J.M. | | 登録日 | 2019-01-30 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Minimally disruptive optical control of protein tyrosine phosphatase 1B.
Nat Commun, 11, 2020
|
|
8E3Q
 
 | | CRYO-EM STRUCTURE OF the human MPSF | | 分子名称: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, ZINC ION, ... | | 著者 | Gutierrez, P.A, Wei, J, Sun, Y, Tong, L. | | 登録日 | 2022-08-17 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Molecular basis for the recognition of the AUUAAA polyadenylation signal by mPSF.
Rna, 28, 2022
|
|
2FI5
 
 | | Crystal structure of a BPTI variant (Cys38->Ser) in complex with trypsin | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | 著者 | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | 登録日 | 2005-12-27 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Functional and structural roles of the Cys14-Cys38 disulfide of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 382, 2008
|
|
8UC6
 
 | | Calpain-7:IST1 Complex | | 分子名称: | Calpain-7, IST1 homolog | | 著者 | Paine, E, Whitby, F.G, Hill, C.P, Sunquist, W.I. | | 登録日 | 2023-09-25 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Elife, 12, 2023
|
|
6H45
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | 分子名称: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Johannsson, S, Neumann, P, Ficner, R. | | 登録日 | 2018-07-20 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-03-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
8C8G
 
 | |
4FZG
 
 | | 20S yeast proteasome in complex with glidobactin | | 分子名称: | Glidobactin, Proteasome component C1, Proteasome component C11, ... | | 著者 | Stein, M, Beck, P, Kaiser, M, Dudler, R, Becker, C.F.W, Groll, M. | | 登録日 | 2012-07-06 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6JC6
 
 | |
3FLP
 
 | |
3FMZ
 
 | |
6UMT
 
 | | High-affinity human PD-1 PD-L2 complex | | 分子名称: | MAGNESIUM ION, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | 著者 | Tang, S, Kim, P.S. | | 登録日 | 2019-10-10 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.986 Å) | | 主引用文献 | A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3T39
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, GLYCEROL, Peptidoglycan recognition protein 1, ... | | 著者 | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2011-07-25 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution
To be Published
|
|
3OS2
 
 | | PFV target capture complex (TCC) at 3.32 A resolution | | 分子名称: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*GP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*CP*TP*CP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | 著者 | Maertens, G.N, Hare, S, Cherepanov, P. | | 登録日 | 2010-09-08 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | The mechanism of retroviral integration from X-ray structures of its key intermediates
Nature, 468, 2010
|
|
