4V47
 
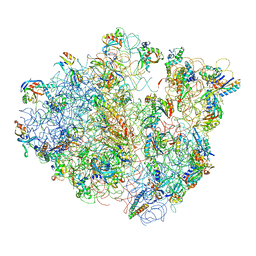 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4V4F
 
 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a RNA molecule containing UAGAU repeats | | Descriptor: | 5'-R(*UP*AP*GP*AP*UP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
4V9S
 
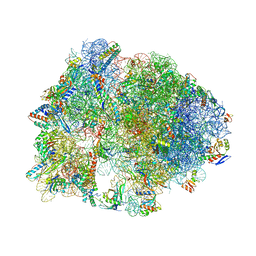 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4US4
 
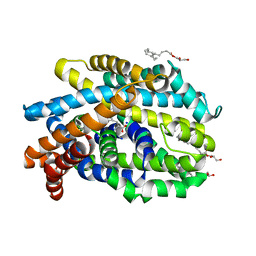 | | Crystal Structure of the Bacterial NSS Member MhsT in an Occluded Inward-Facing State (lipidic cubic phase form) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, SODIUM ION, ... | | Authors: | Malinauskaite, L, Quick, M, Reinhard, L, Lyons, J.A, Yano, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mechanism for Intracellular Release of Na+ by Neurotransmitter/Sodium Symporters
Nat.Struct.Mol.Biol., 21, 2014
|
|
1MOE
 
 | | The three-dimensional structure of an engineered scFv T84.66 dimer or diabody in VL to VH linkage. | | Descriptor: | SULFATE ION, anti-CEA mAb T84.66 | | Authors: | Carmichael, J.A, Power, B.E, Garrett, T.P.J, Yazaki, P.J, Shively, J.E, Raubischek, A.A, Wu, A.M, Hudson, P.J. | | Deposit date: | 2002-09-09 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of an Anti-CEA scFv Diabody Assembled from T84.66 scFvs in VL-to-VH Orientation: Implications for Diabody Flexibility
J.Mol.Biol., 326, 2003
|
|
8XUT
 
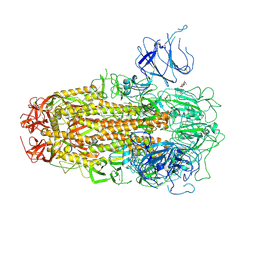 | | XBB.1.5 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P, Mao, X. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
1M0V
 
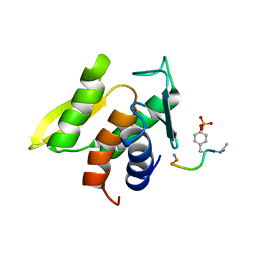 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
8S55
 
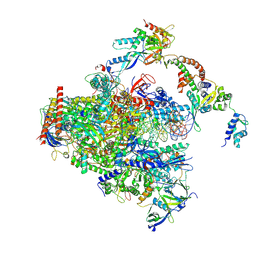 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S51
 
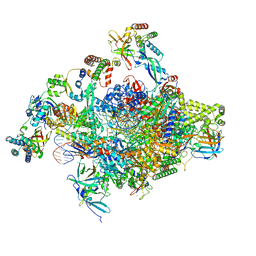 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
4UX8
 
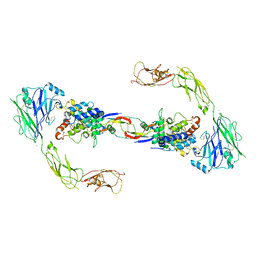 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
6ZGC
 
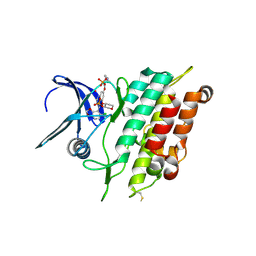 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Saracatinib (AZD0530) | | Descriptor: | Activin receptor type I, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, PHOSPHATE ION, ... | | Authors: | Williams, E.P, Galan Bartual, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Saracatinib is an efficacious clinical candidate for fibrodysplasia ossificans progressiva.
JCI Insight, 6, 2021
|
|
4FR4
 
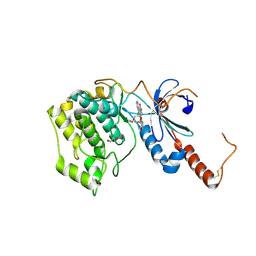 | | Crystal structure of human serine/threonine-protein kinase 32A (YANK1) | | Descriptor: | 1,2-ETHANEDIOL, STAUROSPORINE, Serine/threonine-protein kinase 32A | | Authors: | Chaikuad, A, Elkins, J.M, Krojer, T, Mahajan, P, Goubin, S, Szklarz, M, Tumber, A, Wang, J, Savitsky, P, Shrestha, B, Daga, N, Picaud, S, Fedorov, O, Allerston, C.K, Latwiel, S.V.A, Vollmar, M, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of human serine/threonine-protein kinase 32A (YANK1)
To be Published
|
|
5VM5
 
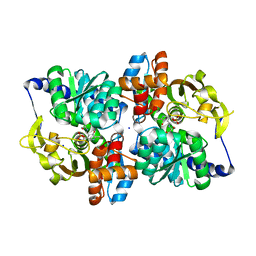 | | Engineered tryptophan synthase b-subunit from Pyrococcus furiosus, PfTrpB2B9, with Ser bound | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, SODIUM ION, Tryptophan synthase beta chain 1, ... | | Authors: | Buller, A.R, van Roye, P. | | Deposit date: | 2017-04-26 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble.
J. Am. Chem. Soc., 140, 2018
|
|
4V9N
 
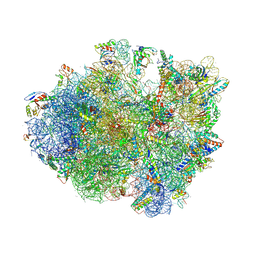 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
8S54
 
 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
4V4M
 
 | | 1.45 Angstrom Structure of STNV coat protein | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Lane, S.W, Dennis, C.A, Lane, C.L, Trinh, C.H, Rizkallah, P.J, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2011-04-28 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Construction and crystal structure of recombinant STNV capsids.
J.Mol.Biol., 413, 2011
|
|
1MJE
 
 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
7REQ
 
 | |
5W3Q
 
 | | L28F E.coli DHFR in complex with NADPH | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Oyen, D, Wright, P.E, Wilson, I.A. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
6ZIX
 
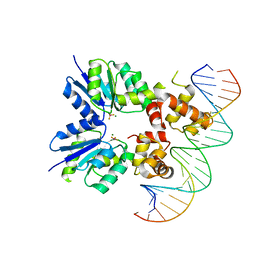 | | Structure of RcsB from Salmonella enterica serovar Typhimurium bound to promoter P1flhDC in the presence of phosphomimetic BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, P1flhDC promoter sequence of 23 bp, ... | | Authors: | Huesa, J, Marina, A, Casino, P. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon.
Nucleic Acids Res., 49, 2021
|
|
5VUZ
 
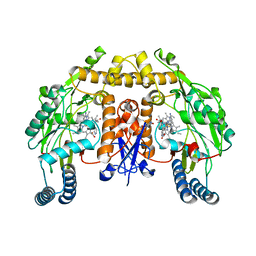 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((3-(Pyridin-3-yl)propyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-(((3-(PYRIDIN-3-YL)PROPYL)AMINO)METHYL)QUINOLIN-2-, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5VUV
 
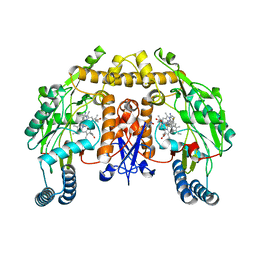 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 7-(((3-Fluorophenyl)amino)methyl)quinolin-2-amine Dihydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{[(3-fluorophenyl)amino]methyl}quinolin-2-amine, Nitric oxide synthase, ... | | Authors: | Huiying, L, Thomas, L.P. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
6QPA
 
 | | Halothiobacillus neapolitanus sulfur oxygenase reductase | | Descriptor: | FE (III) ION, SULFATE ION, Sulfur oxygenase/reductase | | Authors: | Frazao, C, Klezin, A, Poell, U, Veith, A, Ruehl, P, Coelho, R. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple sulfane modifications in active-site cysteine thiols of two sulfur oxygenase reductases and analysis of substrate/product channels
To Be Published
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
