7BOI
 
 | | Bacterial 30S ribosomal subunit assembly complex state F (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8DPO
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-15 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide
To Be Published
|
|
7BOF
 
 | | Bacterial 30S ribosomal subunit assembly complex state I (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
6ZVR
 
 | | C11 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW4
 
 | | C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
8DNT
 
 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
6ZVT
 
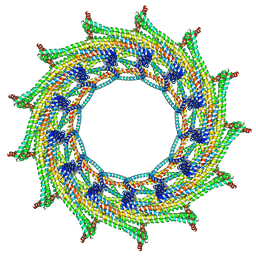 | | C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
4UBK
 
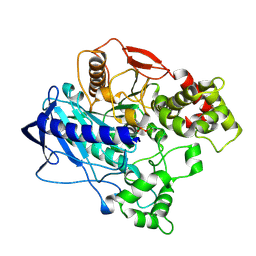 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 7.40 MGy at 100K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
6TXJ
 
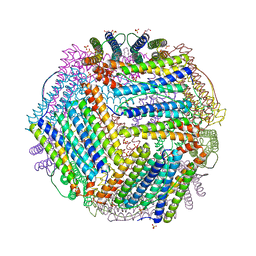 | | Crystal structure of thermotoga maritima A42V E65D Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S, Biela, A.P. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6ZW7
 
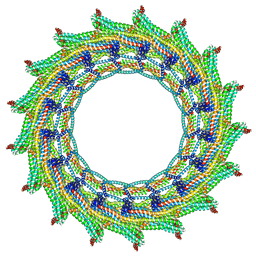 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
4U3Y
 
 | |
4U41
 
 | | MAP4K4 Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U0O
 
 | | Crystal structure of Thermosynechococcus elongatus Lipoyl Synthase 2 complexed with MTA and DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Dinis, P.C, Sandy, J, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
5VWF
 
 | | HLA-B*58:03 presenting LTVQVARVY | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nonamer peptide: LEU-THR-VAL-GLN-VAL-ALA-ARG-VAL-TYR | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2017-05-21 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HLA-B57 micropolymorphism defines the sequence and conformational breadth of the immunopeptidome.
Nat Commun, 9, 2018
|
|
4U1F
 
 | |
7U2B
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (53-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
5ORO
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 3-(4-chlorophenyl)-5,6-dihydroimidazo[2,1-b][1,3]thiazole, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
4UBN
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 1.85 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
5ORR
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 4-[4-(trifluoromethyl)phenyl]-1,2,3-thiadiazol-5-amine, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
5OND
 
 | | RfaH from Escherichia coli in complex with ops DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*TP*AP*GP*TP*C)-3'), Transcription antitermination protein RfaH | | Authors: | Zuber, P.K, Artsimovitch, I, Roesch, P, Knauer, S.H. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand.
Elife, 7, 2018
|
|
5VND
 
 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | Authors: | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | Deposit date: | 2017-04-30 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
