2MW7
 
 | |
2N1F
 
 | | Structure and assembly of the mouse ASC filament by combined NMR spectroscopy and cryo-electron microscopy | | Descriptor: | Apoptosis-associated speck-like protein | | Authors: | Sborgi, L, Ravotti, F, Dandey, V, Dick, M, Mazur, A, Reckel, S, Chami, M, Scherer, S, Bockmann, A, Egelman, E, Stahlberg, H, Broz, P, Meier, B, Hiller, S. | | Deposit date: | 2015-04-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3BSB
 
 | | Crystal Structure of Human Pumilio1 in Complex with CyclinB reverse RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*AP*UP*GP*UP*U)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-23 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
2MRW
 
 | | Solution Structure of MciZ from Bacillus subtilis | | Descriptor: | Cell division factor | | Authors: | Castellen, P, Sforca, M.L, Zeri, A.C.M, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | FtsZ filament capping by MciZ, a developmental regulator of bacterial division.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2KAS
 
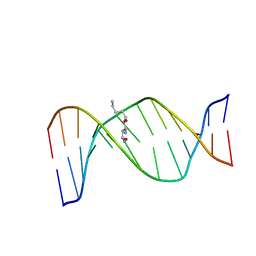 | | HNE-dG adduct mismatched with dA in basic solution | | Descriptor: | (4S)-nonane-1,4-diol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DAP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-11-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational interconversion of the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N(2)-deoxyguanosine adduct when mismatched with deoxyadenosine in DNA
Chem.Res.Toxicol., 22, 2009
|
|
1H70
 
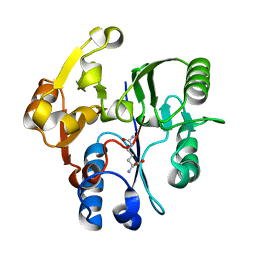 | | DDAH FROM PSEUDOMONAS AERUGINOSA. C249S MUTANT COMPLEXED WITH CITRULLINE | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE | | Authors: | Murray-Rust, J, Leiper, J, McAlister, M, Phelan, J, Tilley, S, Santamaria, J, Vallance, P, McDonald, N. | | Deposit date: | 2001-06-30 | | Release date: | 2001-08-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase.
Nat. Struct. Biol., 8, 2001
|
|
2N4V
 
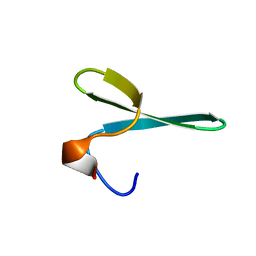 | |
2HZS
 
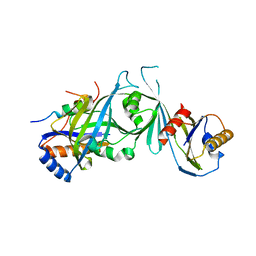 | | Structure of the Mediator head submodule Med8C/18/20 | | Descriptor: | RNA polymerase II mediator complex subunit 18, RNA polymerase II mediator complex subunit 20, RNA polymerase II mediator complex subunit 8 | | Authors: | Lariviere, L, Geiger, S, Hoeppner, S, Rother, S, Straesser, K, Cramer, P. | | Deposit date: | 2006-08-09 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2QQ4
 
 | | Crystal structure of Iron-sulfur cluster biosynthesis protein IscU (TTHA1736) from thermus thermophilus HB8 | | Descriptor: | Iron-sulfur cluster biosynthesis protein IscU, ZINC ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Iron-sulfur cluster biosynthesis protein IscU (TTHA1736) from thermus thermophilus HB8
To be Published
|
|
3U6X
 
 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1SKQ
 
 | | The crystal structure of Sulfolobus solfataricus elongation factor 1-alpha in complex with magnesium and GDP | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vitagliano, L, Ruggiero, A, Masullo, M, Cantiello, P, Arcari, P, Zagari, A. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Sulfolobus solfataricus elongation factor 1alpha in complex with magnesium and GDP.
Biochemistry, 43, 2004
|
|
1SMX
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
2QFD
 
 | | Crystal structure of the regulatory domain of human RIG-I with bound Hg | | Descriptor: | MERCURY (II) ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
1SQN
 
 | | Progesterone Receptor Ligand Binding Domain with bound Norethindrone | | Descriptor: | (14beta,17alpha)-17-ethynyl-17-hydroxyestr-4-en-3-one, progesterone receptor | | Authors: | Williams, S.P, Madauss, K.P, Deng, J.-S, Austin, R.J.H, Lambert, M.H, McLay, I, Pritchard, J, Short, S.A, Stewart, E.L, Uings, I.J. | | Deposit date: | 2004-03-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Progesterone receptor ligand binding pocket flexibility: crystal structures of the norethindrone and mometasone furoate complexes
J.Med.Chem., 47, 2004
|
|
1SQG
 
 | | The crystal structure of the E. coli Fmu apoenzyme at 1.65 A resolution | | Descriptor: | SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
2QKI
 
 | | Human C3c in complex with the inhibitor compstatin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, Complement C3, ... | | Authors: | Janssen, B.J.C, Halff, E.F, Lambris, J.D, Gros, P. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition.
J.Biol.Chem., 282, 2007
|
|
2QLW
 
 | |
1SGG
 
 | | THE SOLUTION STRUCTURE OF SAM DOMAIN FROM THE RECEPTOR TYROSINE KINASE EPHB2, NMR, 10 STRUCTURES | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Smalla, M, Schmieder, P, Kelly, M, Ter Laak, A, Krause, G, Ball, L, Wahl, M, Bork, P, Oschkinat, H. | | Deposit date: | 1999-01-08 | | Release date: | 1999-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites.
Protein Sci., 8, 1999
|
|
2QYH
 
 | | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426 | | Descriptor: | GLYCEROL, Hypothetical conserved protein, GK1056 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-08-15 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426
To be Published
|
|
1VYS
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE W102Y MUTANT AND COMPLEXED WITH PICRIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
2GC0
 
 | |
2GF3
 
 | | Structure of the complex of monomeric sarcosine with its substrate analogue inhibitor 2-furoic acid at 1.3 A resolution. | | Descriptor: | 2-FUROIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, Z, Trickey, P, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-21 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the complex of monomeric sarcosine with its substrate analogue inhibitor 2-furoic acid at 1.3 A resolution.
TO BE PUBLISHED
|
|
2G88
 
 | | MSRECA-dATP COMPLEX | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Krishna, R, Manjunath, G.P, Kumar, P, Surolia, A, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-03-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic identification of an ordered C-terminal domain and a second nucleotide-binding site in RecA: new insights into allostery.
Nucleic Acids Res., 34, 2006
|
|
2GC1
 
 | |
2GCG
 
 | | Ternary Crystal Structure of Human Glyoxylate Reductase/Hydroxypyruvate Reductase | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, Glyoxylate reductase/hydroxypyruvate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Booth, M.P.S, Conners, R, Rumsby, G, Brady, R.L. | | Deposit date: | 2006-03-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate specificity in human glyoxylate reductase/hydroxypyruvate reductase
J.Mol.Biol., 360, 2006
|
|
