5W75
 
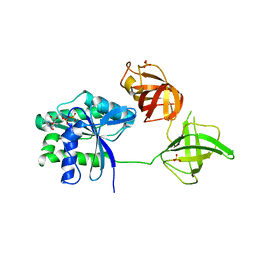 | |
5W76
 
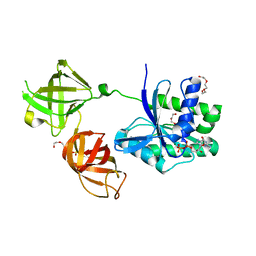 | | Crystal Structure of Reconstructed Bacterial Elongation Factor Node 168 | | Descriptor: | Ancestral Elogation Factor N153, DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ortlund, E.A. | | Deposit date: | 2017-06-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural and Dynamics Comparison of Thermostability in Ancient, Modern, and Consensus Elongation Factor Tus.
Structure, 26, 2018
|
|
4OV7
 
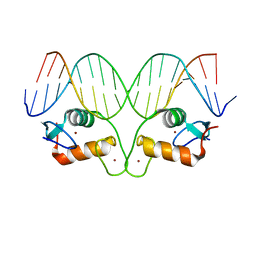 | | Ancestral Steroid Receptor 2 DBD helix mutant - SRE DNA complex | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3', Ancestral Steroid Receptor 2 DBD helix mutant, ... | | Authors: | Ortlund, E.A. | | Deposit date: | 2014-02-20 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evolution of DNA specificity in a transcription factor family produced a new gene regulatory module.
Cell(Cambridge,Mass.), 159, 2014
|
|
5W7Q
 
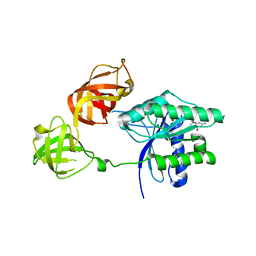 | |
2Q1H
 
 | | Ancestral Corticoid Receptor in Complex with Aldosterone | | Descriptor: | ALDOSTERONE, AncCR, GLYCEROL, ... | | Authors: | Ortlund, E.A, Bridgham, J.T, Redinbo, M.R, Thornton, J.W. | | Deposit date: | 2007-05-24 | | Release date: | 2007-09-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an ancient protein: evolution by conformational epistasis.
Science, 317, 2007
|
|
2Q3Y
 
 | | Ancestral Corticiod Receptor in Complex with DOC | | Descriptor: | Ancestral Corticiod Receptor, DESOXYCORTICOSTERONE, GLYCEROL, ... | | Authors: | Ortlund, E.A, Bridgham, J.T, Redinbo, M.R, Thornton, J.W. | | Deposit date: | 2007-05-30 | | Release date: | 2007-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an ancient protein: evolution by conformational epistasis
Science, 317, 2007
|
|
4M8Z
 
 | |
2Q1V
 
 | | Ancestral corticoid receptor in complex with cortisol | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, AncCR, GLYCEROL | | Authors: | Ortlund, E.A, Redinbo, M.R. | | Deposit date: | 2007-05-25 | | Release date: | 2007-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of an ancient protein: evolution by conformational epistasis.
Science, 317, 2007
|
|
4LTW
 
 | | Ancestral Ketosteroid Receptor-Progesterone-Mifepristone Complex | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Ancestral Steroid Receptor 2, GLYCEROL, ... | | Authors: | Ortlund, E.A, Colucci, J.K. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | X-ray crystal structure of the ancestral 3-ketosteroid receptor-progesterone-mifepristone complex shows mifepristone bound at the coactivator binding interface.
Plos One, 8, 2013
|
|
4FN9
 
 | | X-ray Crystal structure of the Ancestral 3-keto steroid receptor - Progesterone complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Colucci, J.K, Ortlund, E.A, Thornton, J.W. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evolution of minimal specificity and promiscuity in steroid hormone receptors.
PLoS Genet., 8, 2012
|
|
1N2K
 
 | | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, ... | | Authors: | Chruszcz, M, Laidler, P, Monkiewicz, M, Ortlund, E, Lebioda, L, Lewinski, K. | | Deposit date: | 2002-10-23 | | Release date: | 2003-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A.
J.Inorg.Biochem., 96, 2003
|
|
7UOW
 
 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody 034_32 heavy chain, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant evasion from shared neutralizing antibody response.
Structure, 31, 2023
|
|
1N2L
 
 | | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, ... | | Authors: | Chruszcz, M, Laidler, P, Monkiewicz, M, Ortlund, E, Lebioda, L, Lewinski, K. | | Deposit date: | 2002-10-23 | | Release date: | 2003-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A.
J.Inorg.Biochem., 96, 2003
|
|
4TNT
 
 | | Structure of the human mineralocorticoid receptor in complex with DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Mineralocorticoid receptor, ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3938 Å) | | Cite: | Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA.
Plos One, 9, 2014
|
|
8DAF
 
 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
6X6E
 
 | |
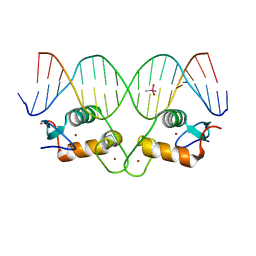
6X6D
 
 | |
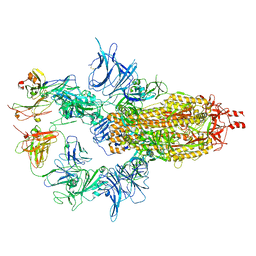
7U0X
 
 | |
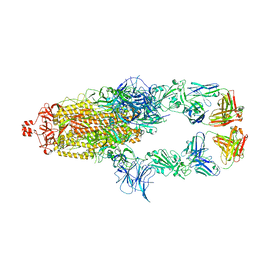
7U0Q
 
 | |
5KRB
 
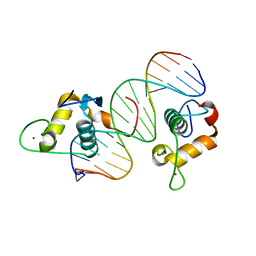 | | GCNF DNA Binding Domain - Oct4 DR0 Complex | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*GP*TP*CP*AP*AP*GP*GP*CP*TP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*GP*CP*CP*TP*TP*GP*AP*CP*CP*TP*CP*T)-3'), Nuclear receptor subfamily 6 group A member 1, ... | | Authors: | Weikum, E.R, Ortlund, E.A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | A Structural Investigation into Oct4 Regulation by Orphan Nuclear Receptors, Germ Cell Nuclear Factor (GCNF), and Liver Receptor Homolog-1 (LRH-1).
J. Mol. Biol., 428, 2016
|
|
6CFN
 
 | |
7U0P
 
 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7UPL
 
 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7JYE
 
 | | Human Liver Receptor Homolog-1 in Complex with 9ChoP and a Fragment of Tif2 | | Descriptor: | 9-[(3~{a}~{R},6~{R},6~{a}~{R})-6-oxidanyl-3-phenyl-3~{a}-(1-phenylethenyl)-4,5,6,6~{a}-tetrahydro-1~{H}-pentalen-2-yl]nonyl 2-(trimethyl-$l^{4}-azanyl)ethyl hydrogen phosphate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
8F8M
 
 | | LRH-1 bound to small molecule Tet and fragment of coactivator Tif2 | | Descriptor: | (1~{R},3~{a}~{R},6~{a}~{R})-4-phenyl-3~{a}-(1-phenylethenyl)-5-[9-(1~{H}-1,2,3,4-tetrazol-5-yl)nonyl]-2,3,6,6~{a}-tetrahydro-1~{H}-pentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isosteric improvements to liver receptor homolog-1 small molecule modulators
To Be Published
|
|
