1TEV
 
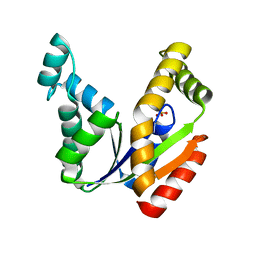 | | Crystal structure of the human UMP/CMP kinase in open conformation | | 分子名称: | SULFATE ION, UMP-CMP kinase | | 著者 | Segura-Pena, D, Sekulic, N, Ort, S, Konrad, M, Lavie, A. | | 登録日 | 2004-05-25 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Substrate-induced Conformational Changes in Human UMP/CMP Kinase.
J.Biol.Chem., 279, 2004
|
|
6CJI
 
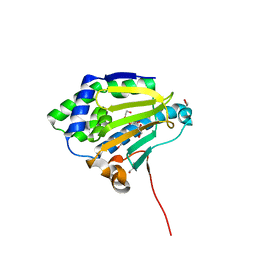 | | Candida albicans Hsp90 nucleotide binding domain | | 分子名称: | 1,2-ETHANEDIOL, Heat shock protein 90 homolog | | 著者 | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, STRUCTURAL GENOMICS CONSORTIUM, S.G.C, Pizarro, J.C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-26 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
1JLC
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | 分子名称: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-16 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLF
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-16 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLE
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE | | 分子名称: | HIV-1 RT, A-CHAIN, B-CHAIN | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-16 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLA
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-651 | | 分子名称: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 RT A-chain, HIV-1 RT B-chain | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-16 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JKH
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-12 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLG
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | 分子名称: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-16 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
4ICC
 
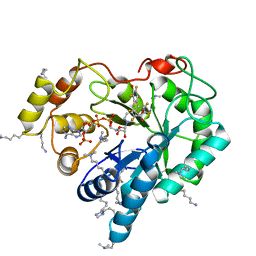 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0064 | | 分子名称: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | 登録日 | 2012-12-10 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IGS
 
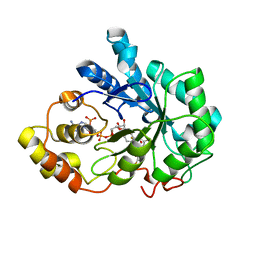 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0064 | | 分子名称: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | 登録日 | 2012-12-18 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2I76
 
 | | Crystal structure of protein TM1727 from Thermotoga maritima | | 分子名称: | Hypothetical protein, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Madegowda, M, Eswaramoorthy, S, Seetharaman, J, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-08-30 | | 公開日 | 2006-10-03 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of hypothetical protein TM1727 from Thermatoga maritima
TO BE PUBLISHED
|
|
2I3O
 
 | |
3EB2
 
 | |
1T3A
 
 | | Crystal structure of Clostridium botulinum neurotoxin type E catalytic domain | | 分子名称: | CHLORIDE ION, ZINC ION, neurotoxin type E | | 著者 | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | 登録日 | 2004-04-26 | | 公開日 | 2004-06-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
1T3C
 
 | | Clostridium botulinum type E catalytic domain E212Q mutant | | 分子名称: | CHLORIDE ION, ZINC ION, neurotoxin type E | | 著者 | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | 登録日 | 2004-04-26 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
2A2Z
 
 | | Crystal Structure of human deoxycytidine kinase in complex with deoxycytidine and uridine diphosphate | | 分子名称: | 2'-DEOXYCYTIDINE, CALCIUM ION, Deoxycytidine kinase, ... | | 著者 | Godsey, M.H, Ort, S, Sabini, E, Konrad, M, Lavie, A. | | 登録日 | 2005-06-23 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Structural basis for the preference of UTP over ATP in human deoxycytidine kinase: illuminating the role of main-chain reorganization.
Biochemistry, 45, 2006
|
|
2A30
 
 | | Crystal structure of human deoxycytidine kinase in complex with deoxycytidine | | 分子名称: | 2'-DEOXYCYTIDINE, CALCIUM ION, Deoxycytidine kinase | | 著者 | Godsey, M.H, Ort, S, Sabini, E, Konrad, M, Lavie, A. | | 登録日 | 2005-06-23 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Structural basis for the preference of UTP over ATP in human deoxycytidine kinase: illuminating the role of main-chain reorganization.
Biochemistry, 45, 2006
|
|
1P60
 
 | | Structure of human dCK complexed with 2'-Deoxycytidine and ADP, Space group C 2 2 21 | | 分子名称: | 2'-DEOXYCYTIDINE, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase | | 著者 | Sabini, E, Ort, S, Monnerjahn, C, Konrad, M, Lavie, A. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
Nat.Struct.Biol., 10, 2003
|
|
1P5Z
 
 | | Structure of human dCK complexed with cytarabine and ADP-MG | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CYTARABINE, Deoxycytidine kinase, ... | | 著者 | Sabini, E, Ort, S, Monnerjahn, C, Konrad, M, Lavie, A. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
Nat.Struct.Biol., 10, 2003
|
|
1P62
 
 | | Structure of human dCK complexed with gemcitabine and ADP-MG | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase, GEMCITABINE, ... | | 著者 | Sabini, E, Ort, S, Monnerjahn, C, Konrad, M, Lavie, A. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
Nat.Struct.Biol., 10, 2003
|
|
1P61
 
 | | Structure of human dCK complexed with 2'-Deoxycytidine and ADP, P 43 21 2 space group | | 分子名称: | 2'-DEOXYCYTIDINE, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase | | 著者 | Sabini, E, Ort, S, Monnerjahn, C, Konrad, M, Lavie, A. | | 登録日 | 2003-04-28 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
Nat.Struct.Biol., 10, 2003
|
|
4RPF
 
 | | Crystal structure of homoserine kinase from Yersinia pestis Nepal516, NYSGRC target 032715 | | 分子名称: | CITRIC ACID, Homoserine kinase | | 著者 | Ptskovsky, Y, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-10-30 | | 公開日 | 2014-11-12 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Homoserine Kinase from Yersinia Pestis Nepal516, Nysgrc Target 032715
To be Published
|
|
4TYM
 
 | | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935 | | 分子名称: | Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-07-08 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935.
To Be Published
|
|
4K9Z
 
 | | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P6222 | | 分子名称: | CHLORIDE ION, Putative thiol-disulfide oxidoreductase | | 著者 | Vetting, M.W, Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R.D, Bonanno, J.B, Armstrong, R.N, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-04-21 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus (target NYSGRC-011676), space group P6222
To be Published
|
|
4JJH
 
 | | Crystal structure of the D1 domain from human Nectin-4 extracellular fragment [PSI-NYSGRC-005624] | | 分子名称: | 1,2-ETHANEDIOL, Poliovirus receptor-related protein 4 | | 著者 | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | 登録日 | 2013-03-07 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the D1 domain of human Nectin-4 extracellular fragment [NYSGRC-005624]
to be published
|
|
