3WD2
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor | | Descriptor: | Chitinase B, DITHIANE DIOL, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD1
 
 | | Serratia marcescens Chitinase B complexed with syn-triazole inhibitor | | Descriptor: | Chitinase B, GLYCEROL, N,N-dibenzyl-N~5~-[N-(methylcarbamoyl)carbamimidoyl]-N~2~-{[5-({[(E)-(quinolin-4-ylmethylidene)amino]oxy}methyl)-1H-1,2,3-triazol-1-yl]acetyl}-L-ornithinamide, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD3
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor | | Descriptor: | Chitinase B, GLYCEROL, SULFATE ION, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD0
 
 | | Serratia marcescens Chitinase B, tetragonal form | | Descriptor: | Chitinase B, DITHIANE DIOL, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3A15
 
 | | Crystal Structure of Substrate-Free Form of Aldoxime Dehydratase (OxdRE) | | Descriptor: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
1AXH
 
 | | ATRACOTOXIN-HVI FROM HADRONYCHE VERSUTA (AUSTRALIAN FUNNEL-WEB SPIDER, NMR, 20 STRUCTURES | | Descriptor: | ATRACOTOXIN-HVI | | Authors: | Fletcher, J.I, O'Donoghue, S.I, Nilges, M, King, G.F. | | Deposit date: | 1996-11-04 | | Release date: | 1997-11-12 | | Last modified: | 2021-02-03 | | Method: | SOLUTION NMR | | Cite: | The structure of a novel insecticidal neurotoxin, omega-atracotoxin-HV1, from the venom of an Australian funnel web spider.
Nat.Struct.Biol., 4, 1997
|
|
1MPH
 
 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | Descriptor: | BETA SPECTRIN | | Authors: | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | Deposit date: | 1997-04-23 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
3P0E
 
 | | Structure of hUPP2 in an active conformation with bound 5-benzylacyclouridine | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, Uridine phosphorylase 2 | | Authors: | Roosild, T.P, Castronovo, S, Villoso, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.
J.Struct.Biol., 176, 2011
|
|
1ORB
 
 | | ACTIVE SITE STRUCTURAL FEATURES FOR CHEMICALLY MODIFIED FORMS OF RHODANESE | | Descriptor: | ACETATE ION, CARBOXYMETHYLATED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1995-07-24 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
2VPL
 
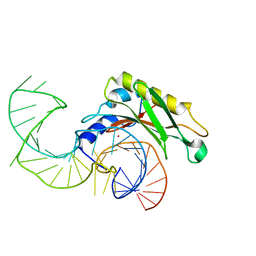 | | The structure of the complex between the first domain of L1 protein from Thermus thermophilus and mRNA from Methanococcus jannaschii | | Descriptor: | 50S RIBOSOMAL PROTEIN L1, FRAGMENT OF MRNA FOR L1-OPERON CONTAINING REGULATOR L1-BINDING SITE, POTASSIUM ION | | Authors: | Kljashtorny, V, Tishchenko, S, Nevskaya, N, Nikonov, S, Garber, M. | | Deposit date: | 2008-03-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Domain II of Thermus thermophilus ribosomal protein L1 hinders recognition of its mRNA.
J. Mol. Biol., 383, 2008
|
|
1I6U
 
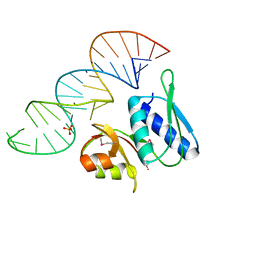 | | RNA-PROTEIN INTERACTIONS: THE CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN S8/RRNA COMPLEX FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 16S RRNA FRAGMENT, 30S RIBOSOMAL PROTEIN S8P, SULFATE ION | | Authors: | Tishchenko, S, Nikulin, A, Fomenkova, N, Nevskaya, N, Nikonov, O, Dumas, P, Moine, H, Ehresmann, B, Ehresmann, C, Piendl, W, Lamzin, V, Garber, M, Nikonov, S. | | Deposit date: | 2001-03-05 | | Release date: | 2001-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii.
J.Mol.Biol., 311, 2001
|
|
1M7L
 
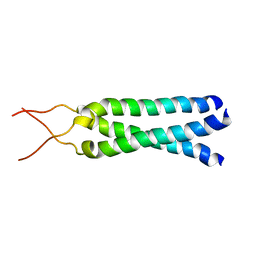 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | Descriptor: | Pulmonary surfactant-associated protein D | | Authors: | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
3INZ
 
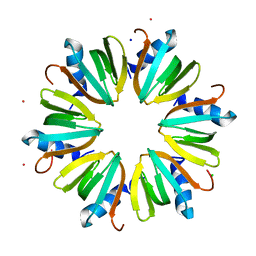 | | H57T Hfq from Pseudomonas aeruginosa | | Descriptor: | CADMIUM ION, CHLORIDE ION, Protein hfq, ... | | Authors: | Moskaleva, O, Melnik, B, Gabdulkhakov, A, Garber, M, Nikonov, S, Stolboushkina, E, Nikulin, A. | | Deposit date: | 2009-08-13 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3TG8
 
 | | Mutant ribosomal protein L1 lacking ala158 from thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, TETRAETHYLENE GLYCOL | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2011-08-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of interdomain mobility in ribosomal L1 proteins.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
4QG3
 
 | | Crystal structure of mutant ribosomal protein G219V TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, BETA-MERCAPTOETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1RZO
 
 | | Agglutinin from Ricinus communis with galactoaza | | Descriptor: | Agglutinin, SULFATE ION, beta-D-galactopyranose | | Authors: | Gabdoulkhakov, A.G, Savochkina, Y, Konareva, N, Krauspenhaar, R, Stoeva, S, Nikonov, S.V, Voelter, W, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2003-12-26 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-function investigation complex of Agglutinin from Ricinus communis with galactoaza
To be Published
|
|
1RYZ
 
 | | Uridine Phosphorylase from Salmonella typhimurium. Crystal Structure at 2.9 A Resolution | | Descriptor: | ACETIC ACID, Uridine phosphorylase | | Authors: | Dontsova, M.V, Gabdoulkhakov, A.G, Lyashenko, A.V, Nikonov, S.V, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2003-12-23 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-functions studies of uridine phosphorylase from Salmonella typhimurium
TO BE PUBLISHED
|
|
4QGB
 
 | | Crystal structure of mutant ribosomal protein G219V TthL1 | | Descriptor: | 50S ribosomal protein L1, ACETATE ION, CHLORIDE ION | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1SJ9
 
 | | Crystal structure of the uridine phosphorylase from Salmonella typhimurium at 2.5A resolution | | Descriptor: | PHOSPHATE ION, Uridine phosphorylase | | Authors: | Dontsova, M, Gabdoulkhakov, A, Morgunova, E, Garber, M, Nikonov, S, Betzel, C, Ealick, S, Mikhailov, A. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preliminary investigation of the three-dimensional structure of Salmonella typhimurium uridine phosphorylase in the crystalline state.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4REO
 
 | | Mutant ribosomal protein l1 from thermus thermophilus with threonine 217 replaced by valine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, GLYCINE, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Tishchenko, S.V, Nikonov, S.V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3A4P
 
 | | human c-MET kinase domain complexed with 6-benzyloxyquinoline inhibitor | | Descriptor: | (2E)-3-{6-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]quinolin-3-yl}-N-methylprop-2-enamide, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Fukami, T.A, Kadono, S, Yamamuro, M, Matsuura, T. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3P0F
 
 | | Structure of hUPP2 in an inactive conformation with bound 5-benzylacyclouridine | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Roosild, T.P, Castronovo, S, Villoso, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.
J.Struct.Biol., 176, 2011
|
|
4PJ3
 
 | | Structural insight into the function and evolution of the spliceosomal helicase Aquarius, Structure of Aquarius in complex with AMPPNP | | Descriptor: | Intron-binding protein aquarius, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | De, I, Bessonov, S, Hofele, R, dos Santos, K.F, Will, C.L, Urlaub, H, Luhrmann, R, Pena, V. | | Deposit date: | 2014-05-11 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2YMX
 
 | | Crystal structure of inhibitory anti-AChE Fab408 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN, GLYCEROL | | Authors: | Bourne, Y, Renault, L, Essono, S, Mondielli, G, Lamourette, P, Bocquet, D, Grassi, J, Marchot, P. | | Deposit date: | 2012-10-10 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Characterization of Monoclonal Antibodies that Inhibit Acetylcholinesterase by Targeting the Peripheral Site and Backdoor Region
Plos One, 8, 2013
|
|
