3ZT4
 
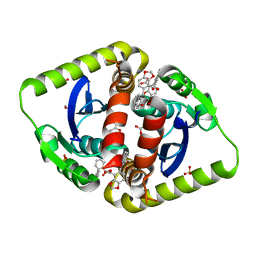 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(2-OXO-2,3-DIHYDRO-1H-INDEN-1-YLIDENE)METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design
Plos One, 7, 2012
|
|
3ZSQ
 
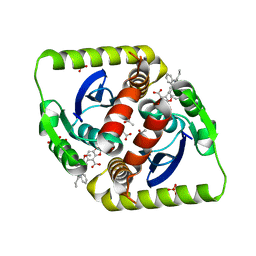 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 5-[[[2-[[(1S)-1-(4-METHOXYPHENYL)BUTYL]CARBAMOYL]PHENYL]METHYL-PROP-2-ENYL-AMINO]METHYL]-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, GLYCEROL, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZT3
 
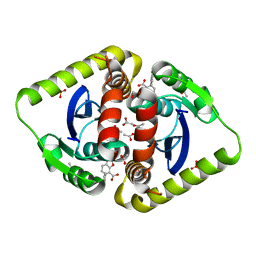 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-{(E)-[(2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YLIDENE]METHYL}-1,3-BENZODIOXOLE-4-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
3ZR7
 
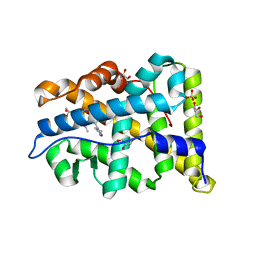 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | 2-CHLORO-N-[[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]METHYL]-1,4-DIMETHYL-1H-PYRAZOLE-4-SULFONAMIDE, GLYCEROL, PROGESTERONE RECEPTOR, ... | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
3ZQW
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-12 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3ZRA
 
 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | N-{(1R)-1-[4-(2-CHLORO-5-FLUOROPYRIDIN-3-YL)PHENYL]ETHYL}-3,5-DIMETHYLISOXAZOLE-4-SULFONAMIDE, PROGESTERONE RECEPTOR, SULFATE ION | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
3ZRB
 
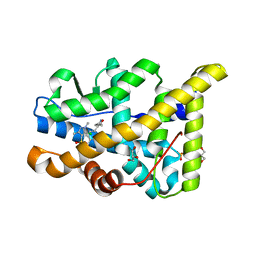 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | (R)-N-[1-[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]ETHYL]-3,5-DIMETHYLISOXAZOLE-4-SULFONAMIDE, 2-CHLORO-N-[[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]METHYL]-1,4-DIMETHYL-1H-PYRAZOLE-4-SULFONAMIDE, GLYCEROL, ... | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, Mcguire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
3ZZZ
 
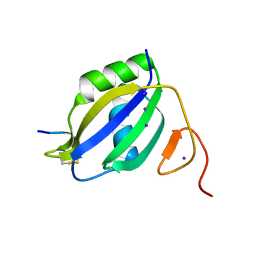 | |
4AHU
 
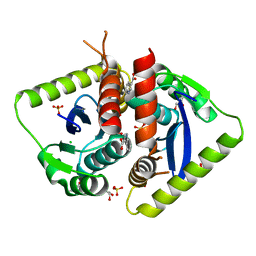 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOLE-3-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4ALG
 
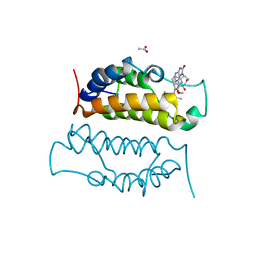 | | N-Terminal Bromodomain of Human BRD2 With IBET-151 | | Descriptor: | 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, ACETATE ION, BROMODOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Lamotte, Y, Donche, F, Bouillot, A, Mirguet, O. | | Deposit date: | 2012-03-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a Novel Series of Bet Family Bromodomain Inhibitors: Binding Mode and Profile of I-Bet151 (Gsk1210151A).
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4A5Z
 
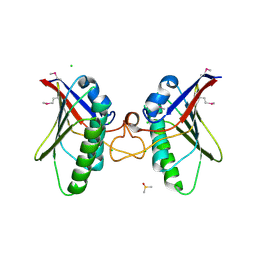 | | Structures of MITD1 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-30 | | Release date: | 2012-11-14 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AQH
 
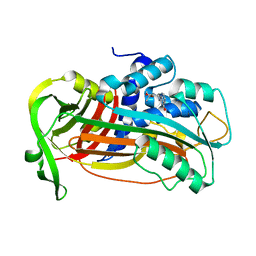 | | Plasminogen activator inhibitor type-1 in complex with the inhibitor AZ3976 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR 1, TERT-BUTYL 3-[(4-OXO-3H-PYRIDO[2,3-D]PYRIMIDIN-2-YL)AMINO]AZETIDINE-1-CARBOXYLATE | | Authors: | Fjellstrom, O, Deinum, J, Sjogren, T, Johansson, C, Geschwindner, S, Nerme, V, Legnehed, A, McPheat, J, Olsson, K, Bodin, C, Gustafsson, D. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a Small Molecule Inhibitor of Plasminogen Activator Inhibitor Type 1 that Accelerates the Transition Into the Latent Conformation
J.Biol.Chem., 288, 2013
|
|
4AQD
 
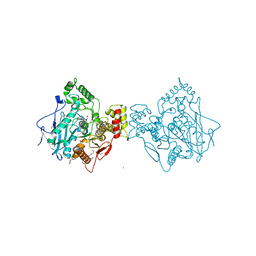 | | Crystal structure of fully glycosylated human butyrylcholinesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Wandhammer, M, Ronco, C, Trovaslet, M, Jean, L, Lockridge, O, Renard, P.Y, Nachon, F. | | Deposit date: | 2012-04-16 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human butyrylcholinesterase produced in insect cells: huprine-based affinity purification and crystal structure.
FEBS J., 279, 2012
|
|
4CB5
 
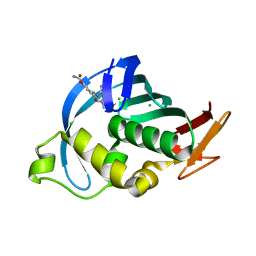 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8f) | | Descriptor: | 9-N-(3-CARBOXY-4-HYDROXYPHENYL)KETOMETHYL-7-N-METHYLGUANINE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CDB
 
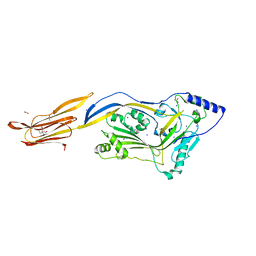 | | Crystal structure of listeriolysin O | | Descriptor: | ACETATE ION, LISTERIOLYSIN O, SODIUM ION, ... | | Authors: | Koester, S, Yildiz, O. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Listeriolysin O Reveals Molecular Details of Oligomerization and Pore Formation
Nat.Commun., 5, 2014
|
|
4C8X
 
 | |
4CB6
 
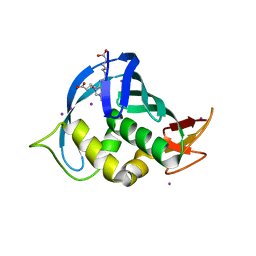 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 11) | | Descriptor: | 2,9-N,N-DI(4-CARBOXYBUTYL)-7-N-METHYLGUANINE, IODIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CBJ
 
 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
4C7N
 
 | | Crystal Structure of the synthetic peptide iM10 in complex with the coiled-coil region of MITF | | Descriptor: | MERCURY (II) ION, MICROPHTHALMIA ASSOCIATED TRANSCRIPTION FACTOR, SYNTHETIC ALPHA-HELIX, ... | | Authors: | Wohlwend, D, Gerhardt, S, Kuekenshoener, T, Einsle, O. | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving Coiled Coil Stability While Maintaining Specificity by a Bacterial Hitchhiker Selection System.
J.Struct.Biol., 186, 2014
|
|
4CB4
 
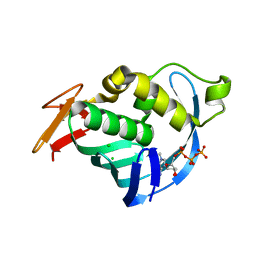 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4C8Y
 
 | |
4BNC
 
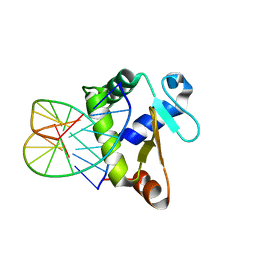 | | Crystal structure of the DNA-binding domain of human ETV1 complexed with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP)-3', HUMAN ETV1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Krojer, T, Chaikuad, A, Vollmar, M, Froese, D.S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2013-05-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4C98
 
 | |
4CG6
 
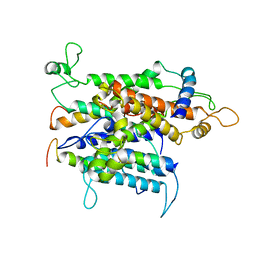 | | Cryo-em of the Sec61-complex bound to the 80s ribosome translating a membrane-inserting substrate | | Descriptor: | PEPTIDE, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT ALPHA ISOFORM 1, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT BETA, ... | | Authors: | Gogala, M, Becker, T, Beatrix, B, Barrio-Garcia, C, Berninghausen, O, Beckmann, R. | | Deposit date: | 2013-11-21 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structures of the Sec61 Complex Engaged in Nascent Peptide Translocation or Membrane Insertion.
Nature, 506, 2014
|
|
4CG7
 
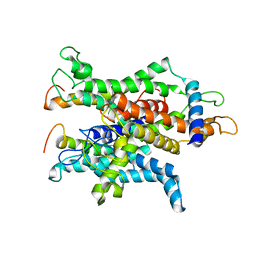 | | Cryo-EM of the Sec61-complex bound to the idle 80S ribosome | | Descriptor: | PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT ALPHA ISOFORM 1, PROTEIN TRANSPORT PROTEIN SEC61 SUBUNIT GAMMA, TRANSPORT PROTEIN SEC61 SUBUNIT BETA | | Authors: | Gogala, M, Becker, T, Beatrix, B, Barrio-Garcia, C, Berninghausen, O, Beckmann, R. | | Deposit date: | 2013-11-21 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structures of the Sec61 Complex Engaged in Nascent Peptide Translocation or Membrane Insertion.
Nature, 506, 2014
|
|
